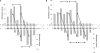Formation of nucleoprotein filaments by mammalian DNA methyltransferase Dnmt3a in complex with regulator Dnmt3L - PubMed (original) (raw)
Formation of nucleoprotein filaments by mammalian DNA methyltransferase Dnmt3a in complex with regulator Dnmt3L
Renata Z Jurkowska et al. Nucleic Acids Res. 2008 Dec.
Abstract
The C-terminal domains of Dnmt3a and Dnmt3L form elongated heterotetramers (3L-3a-3a-3L). Analytical ultracentrifugation confirmed the Dnmt3a-C/3L-C complex exists as a 2:2 heterotetramer in solution. The 3a-3a interface is the DNA-binding site, while both interfaces are essential for AdoMet binding and catalytic activity. Hairpin bisulfite analysis shows correlated methylation of two CG sites in a distance of approximately 8-10 bp in the opposite DNA strands, which corresponds to the geometry of the two active sites in one Dnmt3a-C/3L-C tetramer. Correlated methylation was also observed for two CG sites at similar distances in the same DNA strand, which can be attributed to the binding of two tetramers next to each other. DNA-binding experiments show that Dnmt3a-C/3L-C complexes multimerize on the DNA. Scanning force microscopy demonstrates filament formation rather than binding of single tetramers and shows that protein-DNA filament formation leads to a 1.5-fold shortening of the DNA length.
Figures
Figure 1.
Multimeric state of Dnmt3a-C/3L-C. (A) Model of the Dnmt3a-C/3L-C tetramer, colored in dark and light grey for Dnmt3L-C and Dnmt3a-C, respectively. Modeling of the DNA suggests that the two active sites could methylate two CG sites spaced by ∼10 bp in the opposite DNA strands. (B) Schematic picture of the Dnmt3a-C/3L-C tetramer, the 3a-3L (FF) and 3a–3a (RD) interfaces are indicated. (C) Analytical ultracentrifugation of the Dnmt3a-C/3L-C complex, using concentration corresponding to A280nm of 0.72. Scans were taken every 11.3 min (left panel) and were used to calculate the differential sedimentation coefficient distribution (right panel). (D) SDS-gel showing co-purification of Dnmt3a-C and Dnmt3L-C.
Figure 2.
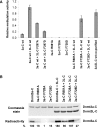
Effects of interface mutations on catalytic activity and cofactor binding. (A) Catalytic activity of the Dnmt3a-C wt and its interface variants analyzed in the absence and presence of Dnmt3L-C by transfer of radioactive methyl groups to a 30-bp oligonucleotide containing one centrally placed CG site. Time courses of DNA methylation were determined over 20 min and used to determine initial slopes. All experiments were carried out at least in triplicate, error bar indicate the standard deviation of the initial slopes. Examples of individual DNA methylation reactions are shown in
Supplementary Figure 1
. (B) AdoMet binding of Dnmt3a-C wt and its mutants in the absence and presence of Dnmt3L-C, as determined by UV-crosslinking of the cofactor to the proteins. After UV crosslink, the proteins were separated on two identical SDS–polyacrylamide gels. The upper panel shows the gel stained with Coomassie, the lower panel shows an autoradiogram indicating the amount of AdoMet bound to the proteins. Densitometric quantification of the signals is shown below the gels in % of the activity observed with wt Dnmt3a-C.
Figure 3.
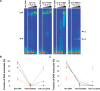
DNA binding of Dnmt3a-C/3L-C and effect of interface mutations on DNA binding. (A) DNA binding of Dnmt3a-C/3L-C wt and mutants determined by EMSA experiment after incubating a 146-bp fluorescently labeled DNA with increasing amounts of protein. It has been shown previously that band 4 observed with wt Dnmt3a-C/3L-C is due to Dnmt3L-C binding to the DNA (19). With Dnmt3a-C F728D and R881A a ladder of bands is also observed in absence of Dnmt3L-C (data not shown). (B and C) Densitometric analysis of the complexes formed by Dnmt3a-C/3L-C at four different enzyme concentrations: 1 µM (blue), 2.5 µM (red), 5 µM (green) and 10 µM (orange). Data are averages from two independent gels, fitted to a fully cooperative (B) and a noncooperative (C) binding model. Error bars represent the deviations between the gels.
Figure 4.
Scanning force microscopy of Dnmt3a-C/3L-C DNA filaments. (A) Dimensions of SFM complexes. In the upper part, a partially occupied DNA molecule is shown. Regions covered with protein are indicated and the length and width of the filaments defined. The height of the filament is defined by the _Z_-axis and encoded in the grayscale of the image. In the lower part, height profiles of sections through a protein/DNA filament (black) and an unoccupied DNA (gray) are shown. The scale bars are 50 nm. (B–E) Examples of SFM complexes: (B) Images showing free DNA. (C–E). Images showing examples of Dnmt3a-C/3L-C DNA filaments with low (C), heavy (D) or full (E) occupancy with protein. In each panel in B–E the frame size is 150 nm. (F) Distribution of DNA length compaction factors of 116 filaments. The distribution of the lengths’ observed for free DNA and fully occupied DNA are shown in
Supplementary Figure 4
.
Figure 5.
Methylation pattern generated by Dnmt3a-C/3L-C. DNA methylation analysis by hairpin bisulfite experiments. Double-stranded oligonucleotide substrates containing either 9 (A) or 12 (B) equally spaced CG sites were methylated by Dnmt3a-C/3L-C complex and the methylation pattern was analyzed by the hairpin bisulfite method. 260 and 169 clones were analyzed for the 9xCG and 12xCG substrates, respectively. The _P_-values for the enrichment of methylation at the particular sites are given in
Supplementary Text 1
. Arrows connect peaks of methylation separated by 8–10 bp in the same or opposite DNA strand.
Similar articles
- Multimerization of the dnmt3a DNA methyltransferase and its functional implications.
Jeltsch A, Jurkowska RZ. Jeltsch A, et al. Prog Mol Biol Transl Sci. 2013;117:445-64. doi: 10.1016/B978-0-12-386931-9.00016-7. Prog Mol Biol Transl Sci. 2013. PMID: 23663978 Review. - Function and disruption of DNA methyltransferase 3a cooperative DNA binding and nucleoprotein filament formation.
Rajavelu A, Jurkowska RZ, Fritz J, Jeltsch A. Rajavelu A, et al. Nucleic Acids Res. 2012 Jan;40(2):569-80. doi: 10.1093/nar/gkr753. Epub 2011 Sep 16. Nucleic Acids Res. 2012. PMID: 21926161 Free PMC article. - Targeted methylation and gene silencing of VEGF-A in human cells by using a designed Dnmt3a-Dnmt3L single-chain fusion protein with increased DNA methylation activity.
Siddique AN, Nunna S, Rajavelu A, Zhang Y, Jurkowska RZ, Reinhardt R, Rots MG, Ragozin S, Jurkowski TP, Jeltsch A. Siddique AN, et al. J Mol Biol. 2013 Feb 8;425(3):479-91. doi: 10.1016/j.jmb.2012.11.038. Epub 2012 Dec 4. J Mol Biol. 2013. PMID: 23220192 - Mechanism of stimulation of catalytic activity of Dnmt3A and Dnmt3B DNA-(cytosine-C5)-methyltransferases by Dnmt3L.
Gowher H, Liebert K, Hermann A, Xu G, Jeltsch A. Gowher H, et al. J Biol Chem. 2005 Apr 8;280(14):13341-8. doi: 10.1074/jbc.M413412200. Epub 2005 Jan 24. J Biol Chem. 2005. PMID: 15671018 - Structure and function of mammalian DNA methyltransferases.
Jurkowska RZ, Jurkowski TP, Jeltsch A. Jurkowska RZ, et al. Chembiochem. 2011 Jan 24;12(2):206-22. doi: 10.1002/cbic.201000195. Epub 2010 Nov 29. Chembiochem. 2011. PMID: 21243710 Review.
Cited by
- Modulation of Dnmt3b function in vitro by interactions with Dnmt3L, Dnmt3a and Dnmt3b splice variants.
Van Emburgh BO, Robertson KD. Van Emburgh BO, et al. Nucleic Acids Res. 2011 Jul;39(12):4984-5002. doi: 10.1093/nar/gkr116. Epub 2011 Mar 4. Nucleic Acids Res. 2011. PMID: 21378119 Free PMC article. - DNA Methyltransferases in Cancer: Biology, Paradox, Aberrations, and Targeted Therapy.
Zhang J, Yang C, Wu C, Cui W, Wang L. Zhang J, et al. Cancers (Basel). 2020 Jul 31;12(8):2123. doi: 10.3390/cancers12082123. Cancers (Basel). 2020. PMID: 32751889 Free PMC article. Review. - DNA Methylation in Lung Cancer: Mechanisms and Associations with Histological Subtypes, Molecular Alterations, and Major Epidemiological Factors.
Hoang PH, Landi MT. Hoang PH, et al. Cancers (Basel). 2022 Feb 15;14(4):961. doi: 10.3390/cancers14040961. Cancers (Basel). 2022. PMID: 35205708 Free PMC article. Review. - Transcription is required for establishment of germline methylation marks at imprinted genes.
Chotalia M, Smallwood SA, Ruf N, Dawson C, Lucifero D, Frontera M, James K, Dean W, Kelsey G. Chotalia M, et al. Genes Dev. 2009 Jan 1;23(1):105-17. doi: 10.1101/gad.495809. Genes Dev. 2009. PMID: 19136628 Free PMC article. - Coordinated chromatin control: structural and functional linkage of DNA and histone methylation.
Cheng X, Blumenthal RM. Cheng X, et al. Biochemistry. 2010 Apr 13;49(14):2999-3008. doi: 10.1021/bi100213t. Biochemistry. 2010. PMID: 20210320 Free PMC article. Review.
References
- Jeltsch A. Beyond Watson and Crick: DNA methylation and molecular enzymology of DNA methyltransferases. Chembiochem. 2002;3:274–293. - PubMed
- Goll MG, Bestor TH. Eukaryotic cytosine methyltransferases. Annu. Rev. Biochem. 2005;74:481–514. - PubMed
- Robertson KD. DNA methylation and human disease. Nat. Rev. Genet. 2005;6:597–610. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases