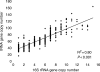rrnDB: documenting the number of rRNA and tRNA genes in bacteria and archaea - PubMed (original) (raw)
. 2009 Jan;37(Database issue):D489-93.
doi: 10.1093/nar/gkn689. Epub 2008 Oct 23.
Affiliations
- PMID: 18948294
- PMCID: PMC2686494
- DOI: 10.1093/nar/gkn689
rrnDB: documenting the number of rRNA and tRNA genes in bacteria and archaea
Zarraz May-Ping Lee et al. Nucleic Acids Res. 2009 Jan.
Abstract
A dramatic exception to the general pattern of single-copy genes in bacterial and archaeal genomes is the presence of 1-15 copies of each ribosomal RNA encoding gene. The original version of the Ribosomal RNA Database (rrnDB) cataloged estimates of the number of 16S rRNA-encoding genes; the database now includes the number of genes encoding each of the rRNAs (5S, 16S and 23S), an internally transcribed spacer region, and the number of tRNA genes. The rrnDB has been used largely by microbiologists to predict the relative rate at which microbial populations respond to favorable growth conditions, and to interpret 16S rRNA-based surveys of microbial communities. To expand the functionality of the rrnDB (http://ribosome.mmg.msu.edu/rrndb/index.php), the search engine has been redesigned to allow database searches based on 16S rRNA gene copy number, specific organisms or taxonomic subsets of organisms. The revamped database also computes average gene copy numbers for any collection of entries selected. Curation tools now permit rapid updates, resulting in an expansion of the database to include data for 785 bacterial and 69 archaeal strains. The rrnDB continues to serve as the authoritative, curated source that documents the phylogenetic distribution of rRNA and tRNA genes in microbial genomes.
Figures
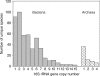
Figure 1.
The number of 16S rRNA genes in bacterial and archaeal genomes. The analysis was performed on 476 bacterial species (gray bars) and 63 archaeal species (checkered bars).
Figure 2.
A screenshot of the result table from the _rrn_DB using ‘search by keyword’ for ‘Vibrio’. The average gene copy number is presented at the end of the table. The dark gray highlighted column indicates that the table is sorted according to the genus name. NA indicates that information for the particular gene is ‘not available’.
Figure 3.
Correlation between the total number of tRNA genes and 16S rRNA genes in bacterial genomes. The data are gathered from sequenced genomes of 590 bacterial species.
References
- Schuwirth BS, Borovinskaya MA, Hau CW, Zhang W, Vila-Sanjurjo A, Holton JM, Cate JH. Structures of the bacterial ribosome at 3.5 A resolution. Science. 2005;310:827–834. - PubMed
- Crosby LD, Criddle CS. Understanding bias in microbial community analysis techniques due to rrn operon copy number heterogeneity. Biotechniques. 2003;34:790–794. 796, 798 passim. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources