Sequence variant classification and reporting: recommendations for improving the interpretation of cancer susceptibility genetic test results - PubMed (original) (raw)
Sequence variant classification and reporting: recommendations for improving the interpretation of cancer susceptibility genetic test results
Sharon E Plon et al. Hum Mutat. 2008 Nov.
Abstract
Genetic testing of cancer susceptibility genes is now widely applied in clinical practice to predict risk of developing cancer. In general, sequence-based testing of germline DNA is used to determine whether an individual carries a change that is clearly likely to disrupt normal gene function. Genetic testing may detect changes that are clearly pathogenic, clearly neutral, or variants of unclear clinical significance. Such variants present a considerable challenge to the diagnostic laboratory and the receiving clinician in terms of interpretation and clear presentation of the implications of the result to the patient. There does not appear to be a consistent approach to interpreting and reporting the clinical significance of variants either among genes or among laboratories. The potential for confusion among clinicians and patients is considerable and misinterpretation may lead to inappropriate clinical consequences. In this article we review the current state of sequence-based genetic testing, describe other standardized reporting systems used in oncology, and propose a standardized classification system for application to sequence-based results for cancer predisposition genes. We suggest a system of five classes of variants based on the degree of likelihood of pathogenicity. Each class is associated with specific recommendations for clinical management of at-risk relatives that will depend on the syndrome. We propose that panels of experts on each cancer predisposition syndrome facilitate the classification scheme and designate appropriate surveillance and cancer management guidelines. The international adoption of a standardized reporting system should improve the clinical utility of sequence-based genetic tests to predict cancer risk.
(c) 2008 Wiley-Liss, Inc.
Figures
Figure 1
The algorithm used for data combination during evaluation of possible carcinogens
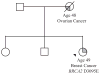
Figure 2
The index case (arrowed) was referred to the genetics service following the diagnosis of a grade 3 breast cancer. Her mother died of a serous papillary ovarian carcinoma aged 48 years. Genetic testing revealed variant D3095E in the BRCA2 gene.
Similar articles
- Assessing pathogenicity: overview of results from the IARC Unclassified Genetic Variants Working Group.
Tavtigian SV, Greenblatt MS, Goldgar DE, Boffetta P; IARC Unclassified Genetic Variants Working Group. Tavtigian SV, et al. Hum Mutat. 2008 Nov;29(11):1261-4. doi: 10.1002/humu.20903. Hum Mutat. 2008. PMID: 18951436 Free PMC article. No abstract available. - Conflicting Interpretation of Genetic Variants and Cancer Risk by Commercial Laboratories as Assessed by the Prospective Registry of Multiplex Testing.
Balmaña J, Digiovanni L, Gaddam P, Walsh MF, Joseph V, Stadler ZK, Nathanson KL, Garber JE, Couch FJ, Offit K, Robson ME, Domchek SM. Balmaña J, et al. J Clin Oncol. 2016 Dec;34(34):4071-4078. doi: 10.1200/JCO.2016.68.4316. Epub 2016 Sep 30. J Clin Oncol. 2016. PMID: 27621404 Free PMC article. - BRCA1 and BRCA2 genetic testing-pitfalls and recommendations for managing variants of uncertain clinical significance.
Eccles DM, Mitchell G, Monteiro AN, Schmutzler R, Couch FJ, Spurdle AB, Gómez-García EB; ENIGMA Clinical Working Group. Eccles DM, et al. Ann Oncol. 2015 Oct;26(10):2057-65. doi: 10.1093/annonc/mdv278. Epub 2015 Jul 7. Ann Oncol. 2015. PMID: 26153499 Free PMC article. Review. - Variant classification changes over time in the clinical molecular diagnostic laboratory setting.
Hahn E, Mighton C, Fisher Y, Wong A, Di Gioacchino V, Watkins N, Mayers J, Bombard Y, Charames GS, Lerner-Ellis J. Hahn E, et al. J Med Genet. 2024 Jul 19;61(8):788-793. doi: 10.1136/jmg-2023-109772. J Med Genet. 2024. PMID: 38806232 - [Genetic diagnosis of cancers? Which cancers should be screened?].
Chompret A. Chompret A. Ann Med Interne (Paris). 2001 Jun;152(4):249-61. Ann Med Interne (Paris). 2001. PMID: 11474373 Review. French.
Cited by
- Reduced penetrance BRCA1 and BRCA2 pathogenic variants in clinical germline genetic testing.
Pal T, Mundt E, Richardson ME, Chao E, Pesaran T, Slavin TP, Couch FJ, Monteiro ANA. Pal T, et al. NPJ Precis Oncol. 2024 Nov 2;8(1):247. doi: 10.1038/s41698-024-00741-4. NPJ Precis Oncol. 2024. PMID: 39488595 Free PMC article. - PIK3CA mutation analysis in circulating tumor cells of patients with hormone receptor positive metastatic breast cancer.
Marino E, Mauro C, Belloni E, Picozzi M, Favalli V, Cassatella MC, Zorzino L, Giacò L, Pelicci PG, Barberis M, Sandri MT, Bernard L. Marino E, et al. Biochem Biophys Rep. 2024 Aug 26;39:101805. doi: 10.1016/j.bbrep.2024.101805. eCollection 2024 Sep. Biochem Biophys Rep. 2024. PMID: 39282094 Free PMC article. - Prevalence and Influence of Genetic Variants on Follow-Up Results in Patients Surviving Thoracic Aortic Therapy.
Ghazy T, Elzanaty N, Lackner HK, Irqsusi M, Rastan AJ, Behrendt CA, Mahlmann A. Ghazy T, et al. J Clin Med. 2024 Sep 5;13(17):5254. doi: 10.3390/jcm13175254. J Clin Med. 2024. PMID: 39274466 Free PMC article. - Assigning credit where it is due: an information content score to capture the clinical value of multiplexed assays of variant effect.
Ranola JMO, Horton C, Pesaran T, Fayer S, Starita LM, Shirts BH. Ranola JMO, et al. BMC Bioinformatics. 2024 Sep 6;25(1):295. doi: 10.1186/s12859-024-05920-5. BMC Bioinformatics. 2024. PMID: 39243022 Free PMC article. - In Silico Prediction of BRCA1 and BRCA2 Variants with Conflicting Clinical Interpretation in a Cohort of Breast Cancer Patients.
Stella S, Vitale SR, Massimino M, Martorana F, Tornabene I, Tomarchio C, Drago M, Pavone G, Gorgone C, Barone C, Bianca S, Manzella L. Stella S, et al. Genes (Basel). 2024 Jul 18;15(7):943. doi: 10.3390/genes15070943. Genes (Basel). 2024. PMID: 39062721 Free PMC article.
References
- Antoniou AC, Cunningham AP, Peto J, Evans DG, Lalloo F, Narod SA, Risch HA, Eyfjord JE, Hopper JL, Southey MC, Olsson H, Johannsson O, Borg A, Passini B, Radice P, Manoukian S, Eccles DM, Tang N, Olah E, nton-Culver H, Warner E, Lubinski J, Gronwald J, Gorski B, Tryggvadottir L, Syrjakoski K, Kallioniemi OP, Eerola H, Nevanlinna H, Pharoah PD, Easton DF. The BOADICEA model of genetic susceptibility to breast and ovarian cancers: updates and extensions. Br J Cancer 2008 - PMC - PubMed
- Balleyguier C, Ayadi S, Van NK, Vanel D, Dromain C, Sigal R. BIRADS classification in mammography. Eur J Radiol. 2007;61:192–194. - PubMed
- Barnetson RA, Cartwright N, van VA, Haq N, Drew K, Farrington S, Williams N, Warner J, Campbell H, Porteous ME, Dunlop MG. Classification of ambiguous mutations in DNA mismatch repair genes identified in a population-based study of colorectal cancer. Hum Mutat. 2008;29:367–374. - PubMed
- Becker F, Nusko G, Welke J, Hahn EG, Mansmann U. Benefit-risk analysis of different risk-related surveillance schedules following colorectal polypectomy. Hepatogastroenterology. 2007;54:2249–2258. - PubMed
- Boerner S, Sneige N. Specimen adequacy and false-negative diagnosis rate in fine-needle aspirates of palpable breast masses. Cancer. 1998;84:344–348. - PubMed
Publication types
MeSH terms
Grants and funding
- R01 HG004064/HG/NHGRI NIH HHS/United States
- MRC_/Medical Research Council/United Kingdom
- U01 CA116167/CA/NCI NIH HHS/United States
- CA116167/CA/NCI NIH HHS/United States
- R01 CA116167/CA/NCI NIH HHS/United States
- R01 CA096536/CA/NCI NIH HHS/United States
- R01 HG004064-02/HG/NHGRI NIH HHS/United States
- 10118/CRUK_/Cancer Research UK/United Kingdom
- R55 CA096536/CA/NCI NIH HHS/United States
- CA 96536/CA/NCI NIH HHS/United States
- HG004064/HG/NHGRI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical