REV1 restrains DNA polymerase zeta to ensure frame fidelity during translesion synthesis of UV photoproducts in vivo - PubMed (original) (raw)
REV1 restrains DNA polymerase zeta to ensure frame fidelity during translesion synthesis of UV photoproducts in vivo
Dávid Szüts et al. Nucleic Acids Res. 2008 Dec.
Abstract
Exposure to ultraviolet light induces a number of forms of damage in DNA, of which (6-4) photoproducts present the most formidable challenge to DNA replication. No single DNA polymerase has been shown to bypass these lesions efficiently in vitro suggesting that the coordinate use of a number of different enzymes is required in vivo. To further understand the mechanisms and control of lesion bypass in vivo, we have devised a plasmid-based system to study the replication of site-specific T-T(6-4) photoproducts in chicken DT40 cells. We show that DNA polymerase zeta is absolutely required for translesion synthesis (TLS) of this lesion, while loss of DNA polymerase eta has no detectable effect. We also show that either the polymerase-binding domain of REV1 or ubiquitinated PCNA is required for the recruitment of Polzeta as the catalytic TLS polymerase. Finally, we demonstrate a previously unappreciated role for REV1 in ensuring bypass synthesis remains in frame with the template. Our data therefore suggest that REV1 not only helps to coordinate the delivery of DNA polymerase zeta to a stalled primer terminus but also restrains its activity to ensure that nucleotides are incorporated in register with the template strand.
Figures
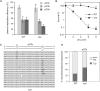
Figure 1.
A system for monitoring lesion bypass in a replicating plasmid. (A) Layout of the pQ1 shuttle plasmid. pUC ori, pUC bacterial replication origin; CMV prom, cytomegalovirus promoter; Gal4-DBD, GAL4 DNA-binding domain; hCdc6, human CDC6 coding sequence; IRES, internal ribosome entry sequence; EGFP, enhanced green fluorescent protein; 5 × Gal4 sites, pentameric Gal4 binding sites. (B) A daily time course of the percentage of wild-type DT40 cells expressing GFP after transfection with 5 µg of pQ1 or a variant lacking the human CDC6 reading frame (pQ1ΔCDC6). Error bars represent 1 SD. (C) A schematic of the staggered arrangement of T–T(6–4) photoproducts in the construct pQTs, with the dinucleotide GC placed opposite each lesion, and 28 bp between the lesion with the possible outcomes of DNA replication over the area. TLS may occur on either the top or the bottom strand, with the most common base insertion shown as (AA). Alternatively, the nascent strand of the sister chromatid may be used as an alternative undamaged template; one possible layout for such a template switching mechanism is illustrated. (D) A schematic representation of the opposing arrangement of T–T(6–4) photoproducts in the construct pQTo and the possible outcomes of DNA replication, only by TLS, over the lesion. (E) Part of a DNA sequencing reaction of the pQTo construct is shown to illustrate the purity of the preparation. The sequencing polymerase stalls at the lesion, and inserts an adenosine opposite the 3′T.
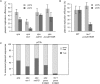
Figure 2.
Lesion bypass in wild-type and xpa cells. (A) Efficiency of replication of shuttle plasmids transfected into WT and xpa cells, and recovered 48 h later. A ligated lesion-free control preparation pQTc as well as two different lesion-containing constructs, pQTs and pQTo were used. These AmpR constructs were co-transfected with the KanR pQT2 plasmid, allowing for the normalization of the number of recovered replicated pQ1-derived constructs against the internal pQ2 control. The average and SEM of 3–5 experiments is shown. (B) Colony survival assay measures the UV light sensitivity of wild-type (WT) and xpa mutant cell lines. Diamonds, WT; squares, xpa. Error bars represent 1 SD. For clarity, only positive error is shown. (C) Example sequences of replicated pQTs plasmids recovered from xpa cells are shown, aligned with a schematic drawing of pQTs. The sequences are sorted, demonstrating TLS on the top strand (inserting mostly AA in the reverse direction, top set), TLS on the bottom strand (middle set), and error-free bypass (bottom set). The proportions are not representative. (D) The proportion of TLS versus error-free bypass in pQTs sequences recovered from wt or xpa cells, shown as percentage of the total. A total of over 80 sequences are shown as a sum of three or more independent experiments.
Figure 3.
REV3 is required for TLS over the T–T(6–4) photoproduct. (A) Replication efficiency of the pQTs shuttle plasmid in xpa, xpa rev3 and xpa polh cells, normalized against an internal pQ2 control as in Figure 2. The average and SEM of 3–5 experiments is shown. (B) Replication efficiency of the pQTo shuttle plasmid in xpa and xpa rev3 cells. (C) The proportion of TLS versus error-free bypass in pQTs sequences recovered from xpa, xpa rev3 and xpa polh cells, shown as a percentage of the total. (D and E) Aligned example sequences from replicated pQTs (D) and pQTo (E) constructs recovered from xpa rev3 cells.
Figure 4.
The dual control of TLS by REV1 and PCNA ubiquitination. (A and B) Replication efficiency of the pQTs and pQTo shuttle plasmids in xpa, xpa rev1, xpa rad18 and xpa pcnaK164R cells (A), and in wt and rev1 pcnaK164R cells (B), normalized against an internal pQ2 control as in Figure 2. The average and SEM of 3–5 experiments is shown. (C) The proportion of TLS versus error-free bypass in pQTs sequences recovered from the indicated cell lines, shown as a percentage of the total.
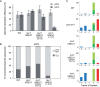
Figure 5.
The pattern of nucleotide incorporation opposite the T–T(6–4) photoproduct. Data for pQTs and pQTo are shown individually. Error-free pQTs sequences are excluded from the analysis. The percentage of each nucleotide incorporated at each position is indicated by the size of the letter of the nucleotide in the column; del, deletion. The incorporation positions indicated are at the 3′T and 5′T of the lesion followed by the next two bases in the template, indicated 1 and 2. Insertions before the + 1 position are indicated in the column ‘ins’, which is also indicated by a shaded grey box. The pQTs plasmids recovered from the xpa rev3 and _rev1 pcna_K164R lines had replicated almost exclusively by an error-free mechanism and generated to few mutant sequences to be plotted, indicated by ‘na’ (not applicable); nd, not done.
Figure 6.
REV1 is required for avoiding frameshifts during TLS. (A) The outcome of TLS in the cell lines indicated. Sequences are classified according to frameshifts (incorrect number of bases inserted opposite the lesion), with deletions shown as <−2, −2 or −1 (blue), the correct two-base insertion as 0 (green), and an extra base as +1 (red). More than one extra base was never observed. The non-frameshifted sequences are further classified, shown as a stacked column, as correct bypass (AA inserted, dark green) or incorrect bypass (light green). In case of pQTs (on the left), the error-free sequences were removed from the analysis; n/a, not applicable. The pQTs samples from the xpa rev3 and _rev1 pcna_K164R cell lines gave almost exclusively error-free results, and therefore could not be plotted, indicated as ‘na’ (not applicable); nd, not done. (B) Aligned example sequences from replicated pQTo constructs recovered from xpa rev1 cells. The different types of frameshift are annotated on the left.
Figure 7.
The C-terminal domain of REV1 is required for its role in restraining Polζ. (A) Replication efficiency of the pQTs and pQTo shuttle plasmids assayed in wt cells and in rev1 mutant cells complemented with full length human REV1 (hREV1), a catalytically inactive point mutant (hREV1[D570AE571A]) and a C-terminal truncation starting at residue 1138 (hREV1[1–1137]). The average and SEM of 3–5 experiments is shown. (B) The proportion of TLS versus error-free bypass in pQTs sequences recovered from the indicated cell lines, shown as a percentage of the total. (C) The outcome of TLS in pQTo in the cell lines indicated, classified according to frameshifts as in Figure 6.
Similar articles
- Analysis of CPD ultraviolet lesion bypass in chicken DT40 cells: polymerase η and PCNA ubiquitylation play identical roles.
Varga A, Marcus AP, Himoto M, Iwai S, Szüts D. Varga A, et al. PLoS One. 2012;7(12):e52472. doi: 10.1371/journal.pone.0052472. Epub 2012 Dec 18. PLoS One. 2012. PMID: 23272247 Free PMC article. - Temporally distinct translesion synthesis pathways for ultraviolet light-induced photoproducts in the mammalian genome.
Temviriyanukul P, van Hees-Stuivenberg S, Delbos F, Jacobs H, de Wind N, Jansen JG. Temviriyanukul P, et al. DNA Repair (Amst). 2012 Jun 1;11(6):550-8. doi: 10.1016/j.dnarep.2012.03.007. Epub 2012 Apr 20. DNA Repair (Amst). 2012. PMID: 22521143 - Mismatch repair protein MSH2 regulates translesion DNA synthesis following exposure of cells to UV radiation.
Lv L, Wang F, Ma X, Yang Y, Wang Z, Liu H, Li X, Liu Z, Zhang T, Huang M, Friedberg EC, Tang TS, Guo C. Lv L, et al. Nucleic Acids Res. 2013 Dec;41(22):10312-22. doi: 10.1093/nar/gkt793. Epub 2013 Sep 12. Nucleic Acids Res. 2013. PMID: 24038355 Free PMC article. - Ubiquitin-dependent regulation of translesion polymerases.
Chun AC, Jin DY. Chun AC, et al. Biochem Soc Trans. 2010 Feb;38(Pt 1):110-5. doi: 10.1042/BST0380110. Biochem Soc Trans. 2010. PMID: 20074045 Review. - Ubiquitination of PCNA and the polymerase switch in human cells.
Kannouche PL, Lehmann AR. Kannouche PL, et al. Cell Cycle. 2004 Aug;3(8):1011-3. Epub 2004 Aug 7. Cell Cycle. 2004. PMID: 15280666 Review.
Cited by
- Regulation of the Rev1-pol ζ complex during bypass of a DNA interstrand cross-link.
Budzowska M, Graham TG, Sobeck A, Waga S, Walter JC. Budzowska M, et al. EMBO J. 2015 Jul 14;34(14):1971-85. doi: 10.15252/embj.201490878. Epub 2015 Jun 12. EMBO J. 2015. PMID: 26071591 Free PMC article. - Y-family DNA polymerase-independent gap-filling translesion synthesis across aristolochic acid-derived adenine adducts in mouse cells.
Hashimoto K, Bonala R, Johnson F, Grollman AP, Moriya M. Hashimoto K, et al. DNA Repair (Amst). 2016 Oct;46:55-60. doi: 10.1016/j.dnarep.2016.07.003. Epub 2016 Jul 29. DNA Repair (Amst). 2016. PMID: 27497692 Free PMC article. - FANCJ coordinates two pathways that maintain epigenetic stability at G-quadruplex DNA.
Sarkies P, Murat P, Phillips LG, Patel KJ, Balasubramanian S, Sale JE. Sarkies P, et al. Nucleic Acids Res. 2012 Feb;40(4):1485-98. doi: 10.1093/nar/gkr868. Epub 2011 Oct 22. Nucleic Acids Res. 2012. PMID: 22021381 Free PMC article. - DNA polymerases and cancer.
Lange SS, Takata K, Wood RD. Lange SS, et al. Nat Rev Cancer. 2011 Feb;11(2):96-110. doi: 10.1038/nrc2998. Nat Rev Cancer. 2011. PMID: 21258395 Free PMC article. Review. - Histone H3.3 is required to maintain replication fork progression after UV damage.
Frey A, Listovsky T, Guilbaud G, Sarkies P, Sale JE. Frey A, et al. Curr Biol. 2014 Sep 22;24(18):2195-2201. doi: 10.1016/j.cub.2014.07.077. Epub 2014 Sep 4. Curr Biol. 2014. PMID: 25201682 Free PMC article.
References
- Prakash S, Johnson RE, Prakash L. Eukaryotic translesion synthesis DNA polymerases: specificity of structure and function. Annu. Rev. Biochem. 2005;74:317–353. - PubMed
- Hoege C, Pfander B, Moldovan GL, Pyrowolakis G, Jentsch S. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature. 2002;419:135–141. - PubMed
- Kannouche PL, Wing J, Lehmann AR. Interaction of human DNA polymerase η with monoubiquitinated PCNA: a possible mechanism for the polymerase switch in response to DNA damage. Mol. Cell. 2004;14:491–500. - PubMed
- Bienko M, Green CM, Crosetto N, Rudolf F, Zapart G, Coull B, Kannouche P, Wider G, Peter M, Lehmann AR, et al. Ubiquitin-binding domains in Y-family polymerases regulate translesion synthesis. Science. 2005;310:1821–1824. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous