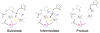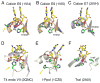An equivalent metal ion in one- and two-metal-ion catalysis - PubMed (original) (raw)
. 2008 Nov;15(11):1228-31.
doi: 10.1038/nsmb.1502. Epub 2008 Oct 26.
Affiliations
- PMID: 18953336
- PMCID: PMC2597392
- DOI: 10.1038/nsmb.1502
An equivalent metal ion in one- and two-metal-ion catalysis
Wei Yang. Nat Struct Mol Biol. 2008 Nov.
Abstract
Nucleotidyl-transfer enzymes, which synthesize, degrade and rearrange DNA and RNA, often depend on metal ions for catalysis. All DNA and RNA polymerases, MutH-like or RNase H-like nucleases and recombinases, and group I introns seem to require two divalent cations to form a complete active site. The two-metal-ion mechanism has been proposed to orient the substrate, facilitate acid-base catalysis and allow catalytic specificity to exceed substrate binding specificity attributable to the stringent metal-ion (Mg2+ in particular) coordination. Not all nucleotidyl-transfer enzymes use two metal ions for catalysis, however. The betabetaalpha-Me and HUH nucleases depend on a single metal ion in the active site for the catalysis. All of these one- and two metal ion-dependent enzymes generate 5'-phosphate and 3'-OH products. Structural and mechanistic comparisons show that these seemingly unrelated nucleotidyl-transferases share a functionally equivalent metal ion.
Figures
Figure 1. A diagram of two-metal ion catalysis by RNase H
Coordination of metal ions A and B is indicated by the dashed lines. The Asp conserved in all nucleotidyl-transfer enzymes using two-metal ions is highlighted in pink. This Asp is replaced by a backbone phosphate in group I introns. The oxygen atoms from the protein and scissile phosphate that chelate metal ion B are highlighted in pink, and the attacking nucleophile is highlighted in blue.
Figure 2. Examples of one-metal ion-dependent nucleases
(A–E) The ββα-Me family. (F) The flipped ββα-Me family. The active site residues are shown in green(C)/blue(N)/red(O) and the DNA substrate centered on the scissile phosphate in yellow(C)/blue/red/orange(P) stick models. The Na+, Mg2+ and Zn2+ in the active site are shown as color-coded spheres. Coordination of metal ion is represented by pink dashed lines. The mobile metal-ion ligand is indicated by a yellow arrow. When determined, the nucleophile water is shown as a red sphere. The adjoining two active site residues in the ββα-Me nucleases are highlighted by the orange ovals. Mutations of the active site residues, which enable crystallization of enzyme-substrate complexes are labeled.
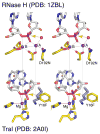
Figure 3. Comparison of the active site of RNase H and TraI in stereoview
The nucleic acid is shown in light grey, and the protein in yellow with the N (blue), O (red), P (meganta) highlighted. The Mg2+ ions are shown as purple spheres, and their coordination by macromolecules is indicated by dashed pink lines. The active site mutations (D192N and Y16F) that prevent the chemistry are labeled. Alignments of the metal ions (or Lys) between the two enzymes are shown by the semi-transparent grey lines. The orange arrows represent the nucleophilic attack.
Similar articles
- Metal-ion-dependent catalysis and specificity of CCA-adding enzymes: a comparison of two classes.
Hou YM, Gu SQ, Zhou H, Ingerman L. Hou YM, et al. Biochemistry. 2005 Sep 27;44(38):12849-59. doi: 10.1021/bi0509402. Biochemistry. 2005. PMID: 16171400 - Crystal structures of RNase H bound to an RNA/DNA hybrid: substrate specificity and metal-dependent catalysis.
Nowotny M, Gaidamakov SA, Crouch RJ, Yang W. Nowotny M, et al. Cell. 2005 Jul 1;121(7):1005-16. doi: 10.1016/j.cell.2005.04.024. Cell. 2005. PMID: 15989951 - A combined experimental and theoretical study of divalent metal ion selectivity and function in proteins: application to E. coli ribonuclease H1.
Babu CS, Dudev T, Casareno R, Cowan JA, Lim C. Babu CS, et al. J Am Chem Soc. 2003 Aug 6;125(31):9318-28. doi: 10.1021/ja034956w. J Am Chem Soc. 2003. PMID: 12889961 - Catalytic metal ions and enzymatic processing of DNA and RNA.
Palermo G, Cavalli A, Klein ML, Alfonso-Prieto M, Dal Peraro M, De Vivo M. Palermo G, et al. Acc Chem Res. 2015 Feb 17;48(2):220-8. doi: 10.1021/ar500314j. Epub 2015 Jan 15. Acc Chem Res. 2015. PMID: 25590654 Review. - Multiple roles of metal ions in large ribozymes.
Donghi D, Schnabl J. Donghi D, et al. Met Ions Life Sci. 2011;9:197-234. doi: 10.1039/9781849732512-00197. Met Ions Life Sci. 2011. PMID: 22010273 Review.
Cited by
- Observing one-divalent-metal-ion-dependent and histidine-promoted His-Me family I-PpoI nuclease catalysis in crystallo.
Chang C, Zhou G, Gao Y. Chang C, et al. Elife. 2024 Aug 14;13:RP99960. doi: 10.7554/eLife.99960. Elife. 2024. PMID: 39141555 Free PMC article. - Recent Updates of the CRISPR/Cas9 Genome Editing System: Novel Approaches to Regulate Its Spatiotemporal Control by Genetic and Physicochemical Strategies.
Allemailem KS, Almatroudi A, Rahmani AH, Alrumaihi F, Alradhi AE, Alsubaiyel AM, Algahtani M, Almousa RM, Mahzari A, Sindi AAA, Dobie G, Khan AA. Allemailem KS, et al. Int J Nanomedicine. 2024 Jun 6;19:5335-5363. doi: 10.2147/IJN.S455574. eCollection 2024. Int J Nanomedicine. 2024. PMID: 38859956 Free PMC article. Review. - Observing one-divalent-metal-ion dependent and histidine-promoted His-Me family I-PpoI nuclease catalysis in crystallo.
Chang C, Zhou G, Gao Y. Chang C, et al. bioRxiv [Preprint]. 2024 Jul 11:2024.05.02.592236. doi: 10.1101/2024.05.02.592236. bioRxiv. 2024. PMID: 38746211 Free PMC article. Updated. Preprint. - Dual-wield NTPases: A novel protein family mined from AlphaFold DB.
Sakuma K, Koike R, Ota M. Sakuma K, et al. Protein Sci. 2024 Apr;33(4):e4934. doi: 10.1002/pro.4934. Protein Sci. 2024. PMID: 38501460 Free PMC article. - Coupled catalytic states and the role of metal coordination in Cas9.
Das A, Rai J, Roth MO, Shu Y, Medina ML, Barakat MR, Li H. Das A, et al. Nat Catal. 2023 Oct;6(10):969-977. doi: 10.1038/s41929-023-01031-1. Epub 2023 Oct 2. Nat Catal. 2023. PMID: 38348449 Free PMC article.
References
- Pelletier H, Sawaya MR, Kumar A, Wilson SH, Kraut J. Structures of ternary complexes of rat DNA polymerase beta, a DNA template-primer, and ddCTP. Science. 1994;264:1891–903. - PubMed
- Doublie S, Tabor S, Long AM, Richardson CC, Ellenberger T. Crystal structure of a bacteriophage T7 DNA replication complex at 2.2 A resolution. Nature. 1998;391:251–8. - PubMed
- Steitz TA. A mechanism for all polymerases. Nature. 1998;391:231–2. - PubMed
- Lee JY, et al. MutH complexed with Hemi- and unmethylated DNAs: coupling base recognition and DNA cleavage. Mol Cell. 2005;20:155–66. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources