Nature, nurture, or chance: stochastic gene expression and its consequences - PubMed (original) (raw)
Review
Nature, nurture, or chance: stochastic gene expression and its consequences
Arjun Raj et al. Cell. 2008.
Abstract
Gene expression is a fundamentally stochastic process, with randomness in transcription and translation leading to cell-to-cell variations in mRNA and protein levels. This variation appears in organisms ranging from microbes to metazoans, and its characteristics depend both on the biophysical parameters governing gene expression and on gene network structure. Stochastic gene expression has important consequences for cellular function, being beneficial in some contexts and harmful in others. These situations include the stress response, metabolism, development, the cell cycle, circadian rhythms, and aging.
Figures
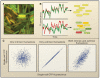
Figure 1. Intrinsic and extrinsic contributions to noise in gene expression
A) A fluorescence image of individual E. coli displaying marked cell-to-cell variability in the expression of two identically regulated fluorescent proteins. B) Schematic depiction of the temporal behaviors of extrinsic noise (upper) and intrinsic noise (lower). C) Expected cell-to-cell variations when fluctuations are intrinsic, extrinsic or both. (A and B adapted from Elowitz et al., 2002).
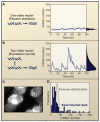
Figure 2. Noise in prokaryotic gene expression depends on the rates of transcription and translation
A) When the transcription rate is high, variability in protein levels is low, but B) when the transcription rate is lowered and the translation rate is raised, gene expression is far noisier, even at the same mean, as shown in Ozbudak et al. (2002).
Figure 3. The Contribution of Transcriptional Bursts to Cell-to-Cell Variability
A) Transcription without bursts with a relatively small amount of noise. B) Bursts in transcription can cause significantly higher variability, even when producing the same mean number of transcripts. C) In situ detection of individual mRNA molecules reveals large cell-to-cell variability in mammalian cells. D) Experimental histogram of mRNA numbers. The grey dashed line depicts the theoretical distribution one would expect in the absence of transcriptional bursts. (C and D adapted from Raj et al., 2006)
Similar articles
- Teasing apart translational and transcriptional components of stochastic variations in eukaryotic gene expression.
Salari R, Wojtowicz D, Zheng J, Levens D, Pilpel Y, Przytycka TM. Salari R, et al. PLoS Comput Biol. 2012;8(8):e1002644. doi: 10.1371/journal.pcbi.1002644. Epub 2012 Aug 30. PLoS Comput Biol. 2012. PMID: 22956896 Free PMC article. - Noise in eukaryotic gene expression.
Blake WJ, KAErn M, Cantor CR, Collins JJ. Blake WJ, et al. Nature. 2003 Apr 10;422(6932):633-7. doi: 10.1038/nature01546. Nature. 2003. PMID: 12687005 - Transcriptional stochasticity in gene expression.
Lipniacki T, Paszek P, Marciniak-Czochra A, Brasier AR, Kimmel M. Lipniacki T, et al. J Theor Biol. 2006 Jan 21;238(2):348-67. doi: 10.1016/j.jtbi.2005.05.032. Epub 2005 Jul 21. J Theor Biol. 2006. PMID: 16039671 - Stochasticity in gene expression: from theories to phenotypes.
Kaern M, Elston TC, Blake WJ, Collins JJ. Kaern M, et al. Nat Rev Genet. 2005 Jun;6(6):451-64. doi: 10.1038/nrg1615. Nat Rev Genet. 2005. PMID: 15883588 Review. - Stochastic models for circadian rhythms: effect of molecular noise on periodic and chaotic behaviour.
Gonze D, Halloy J, Leloup JC, Goldbeter A. Gonze D, et al. C R Biol. 2003 Feb;326(2):189-203. doi: 10.1016/s1631-0691(03)00016-7. C R Biol. 2003. PMID: 12754937 Review.
Cited by
- Method of moments framework for differential expression analysis of single-cell RNA sequencing data.
Kim MC, Gate R, Lee DS, Tolopko A, Lu A, Gordon E, Shifrut E, Garcia-Nieto PE, Marson A, Ntranos V, Ye CJ. Kim MC, et al. Cell. 2024 Oct 31;187(22):6393-6410.e16. doi: 10.1016/j.cell.2024.09.044. Epub 2024 Oct 24. Cell. 2024. PMID: 39454576 - Cell cycle expression heterogeneity predicts degree of differentiation.
Noller K, Cahan P. Noller K, et al. Brief Bioinform. 2024 Sep 23;25(6):bbae536. doi: 10.1093/bib/bbae536. Brief Bioinform. 2024. PMID: 39446193 Free PMC article. - Transcription-dependent mobility of single genes and genome-wide motions in live human cells.
Chu FY, Clavijo AS, Lee S, Zidovska A. Chu FY, et al. Nat Commun. 2024 Oct 22;15(1):8879. doi: 10.1038/s41467-024-51149-4. Nat Commun. 2024. PMID: 39438437 Free PMC article. - MONITTR allows real-time imaging of transcription and endogenous proteins in C. elegans.
Liu X, Chang Z, Sun P, Cao B, Wang Y, Fang J, Pei Y, Chen B, Zou W. Liu X, et al. J Cell Biol. 2025 Jan 6;224(1):e202403198. doi: 10.1083/jcb.202403198. Epub 2024 Oct 14. J Cell Biol. 2025. PMID: 39400293 - A differentiable Gillespie algorithm for simulating chemical kinetics, parameter estimation, and designing synthetic biological circuits.
Rijal K, Mehta P. Rijal K, et al. ArXiv [Preprint]. 2024 Sep 25:arXiv:2407.04865v2. ArXiv. 2024. PMID: 39398212 Free PMC article. Preprint.
References
- Acar M, Becskei A, van Oudenaarden A. Enhancement of cellular memory by reducing stochastic transitions. Nature. 2005;435:228–232. - PubMed
- Acar M, Mettetal JT, van Oudenaarden A. Stochastic switching as a survival strategy in fluctuating environments. Nat Genet. 2008;40:471–475. - PubMed
- Austin DW, Allen MS, McCollum JM, Dar RD, Wilgus JR, Sayler GS, Samatova NF, Cox CD, Simpson ML. Gene network shaping of inherent noise spectra. Nature. 2006;439:608–611. - PubMed
- Bahar R, Hartmann CH, Rodriguez KA, Denny AD, Busuttil RA, Dolle ME, Calder RB, Chisholm GB, Pollock BH, Klein CA, Vijg J. Increased cell-to-cell variation in gene expression in ageing mouse heart. Nature. 2006;441:1011–1014. - PubMed
Publication types
MeSH terms
Grants and funding
- R01-GM077183/GM/NIGMS NIH HHS/United States
- R01 GM077183/GM/NIGMS NIH HHS/United States
- R01 GM068957/GM/NIGMS NIH HHS/United States
- R01 GM077183-03/GM/NIGMS NIH HHS/United States
- R01-GM068957/GM/NIGMS NIH HHS/United States
- R01 GM068957-07/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources