Analyses and comparison of accuracy of different genotype imputation methods - PubMed (original) (raw)
Comparative Study
Analyses and comparison of accuracy of different genotype imputation methods
Yu-Fang Pei et al. PLoS One. 2008.
Abstract
The power of genetic association analyses is often compromised by missing genotypic data which contributes to lack of significant findings, e.g., in in silico replication studies. One solution is to impute untyped SNPs from typed flanking markers, based on known linkage disequilibrium (LD) relationships. Several imputation methods are available and their usefulness in association studies has been demonstrated, but factors affecting their relative performance in accuracy have not been systematically investigated. Therefore, we investigated and compared the performance of five popular genotype imputation methods, MACH, IMPUTE, fastPHASE, PLINK and Beagle, to assess and compare the effects of factors that affect imputation accuracy rates (ARs). Our results showed that a stronger LD and a lower MAF for an untyped marker produced better ARs for all the five methods. We also observed that a greater number of haplotypes in the reference sample resulted in higher ARs for MACH, IMPUTE, PLINK and Beagle, but had little influence on the ARs for fastPHASE. In general, MACH and IMPUTE produced similar results and these two methods consistently outperformed fastPHASE, PLINK and Beagle. Our study is helpful in guiding application of imputation methods in association analyses when genotype data are missing.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
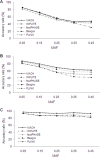
Figure 1. Effects of LD level on accuracy rates.
The results are based on 90 reference haplotypes and a medium marker density (one SNP per 6 kb).
Figure 2. Effects of MAF of untyped SNPs on accuracy rates.
The results are based on 90 reference haplotypes and the medium marker density (1 SNP per 6 kb). (a) Low LD level; (b) Medium LD level; (c) High LD level.
Figure 3. Effects of marker density on accuracy rates.
The results are based on 90 reference haplotypes at the medium LD level. X-axis represents marker density: low marker density: one SNP per 10 kb; medium marker density: one SNP per 6 kb and high marker density: one SNP per 3 kb.
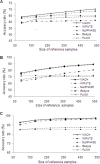
Figure 4. Effects of sample size of reference samples on accuracy rates under various conditions.
(a) Low LD level and high marker density (one SNP per 3 kb); (b) Medium LD level and medium marker density (one SNP per 6 kb); (c) High LD level and low marker density (one SNP per 10 kb).
Figure 5. Performance of the imputation methods under various conditions using real data sets.
Each label along x-axis represents a specific combination of LD level and marker density. Within each label, “L”, “M”, and “H” refer to, respectively, low, medium and high LD level when they are the first letter or marker density when they are the second letter.
Figure 6. Effects of MAF of untyped SNPs on accuracy rates in real datasets.
The results are based on the medium marker density (1 SNP per 6 kb). (a) Low LD level; (b) Medium LD level; (c) High LD level.
Similar articles
- Accuracy of genome-wide imputation of untyped markers and impacts on statistical power for association studies.
Hao K, Chudin E, McElwee J, Schadt EE. Hao K, et al. BMC Genet. 2009 Jun 16;10:27. doi: 10.1186/1471-2156-10-27. BMC Genet. 2009. PMID: 19531258 Free PMC article. - Analyses and comparison of imputation-based association methods.
Pei YF, Zhang L, Li J, Deng HW. Pei YF, et al. PLoS One. 2010 May 26;5(5):e10827. doi: 10.1371/journal.pone.0010827. PLoS One. 2010. PMID: 20520814 Free PMC article. - A comprehensive evaluation of SNP genotype imputation.
Nothnagel M, Ellinghaus D, Schreiber S, Krawczak M, Franke A. Nothnagel M, et al. Hum Genet. 2009 Mar;125(2):163-71. doi: 10.1007/s00439-008-0606-5. Epub 2008 Dec 17. Hum Genet. 2009. PMID: 19089453 - Genotype imputation in genome-wide association studies.
Porcu E, Sanna S, Fuchsberger C, Fritsche LG. Porcu E, et al. Curr Protoc Hum Genet. 2013 Jul;Chapter 1:Unit 1.25. doi: 10.1002/0471142905.hg0125s78. Curr Protoc Hum Genet. 2013. PMID: 23853078 Review. - Genotype Imputation from Large Reference Panels.
Das S, Abecasis GR, Browning BL. Das S, et al. Annu Rev Genomics Hum Genet. 2018 Aug 31;19:73-96. doi: 10.1146/annurev-genom-083117-021602. Epub 2018 May 23. Annu Rev Genomics Hum Genet. 2018. PMID: 29799802 Review.
Cited by
- Prospective evaluation of B-type natriuretic peptide concentrations and the risk of type 2 diabetes in women.
Everett BM, Cook NR, Chasman DI, Magnone MC, Bobadilla M, Rifai N, Ridker PM, Pradhan AD. Everett BM, et al. Clin Chem. 2013 Mar;59(3):557-65. doi: 10.1373/clinchem.2012.194167. Epub 2013 Jan 3. Clin Chem. 2013. PMID: 23288489 Free PMC article. - Evaluation of imputation accuracy using the combination of two high-density panels in Nelore beef cattle.
Bernardes PA, Nascimento GBD, Savegnago RP, Buzanskas ME, Watanabe RN, de Almeida Regitano LC, Coutinho LL, Gondro C, Munari DP. Bernardes PA, et al. Sci Rep. 2019 Nov 29;9(1):17920. doi: 10.1038/s41598-019-54382-w. Sci Rep. 2019. PMID: 31784673 Free PMC article. - Accuracy of imputation to whole-genome sequence data in Holstein Friesian cattle.
van Binsbergen R, Bink MC, Calus MP, van Eeuwijk FA, Hayes BJ, Hulsegge I, Veerkamp RF. van Binsbergen R, et al. Genet Sel Evol. 2014 Jul 15;46(1):41. doi: 10.1186/1297-9686-46-41. Genet Sel Evol. 2014. PMID: 25022768 Free PMC article. - Comparison of the performance of two commercial genome-wide association study genotyping platforms in Han Chinese samples.
Jiang L, Willner D, Danoy P, Xu H, Brown MA. Jiang L, et al. G3 (Bethesda). 2013 Jan;3(1):23-9. doi: 10.1534/g3.112.004069. Epub 2013 Jan 1. G3 (Bethesda). 2013. PMID: 23316436 Free PMC article. - Comparison of different imputation methods from low- to high-density panels using Chinese Holstein cattle.
Weng Z, Zhang Z, Zhang Q, Fu W, He S, Ding X. Weng Z, et al. Animal. 2013 May;7(5):729-35. doi: 10.1017/S1751731112002224. Epub 2012 Dec 11. Animal. 2013. PMID: 23228675 Free PMC article.
References
- de Bakker PI, Yelensky R, Pe'er I, Gabriel SB, Daly MJ, et al. Efficiency and power in genetic association studies. Nat Genet. 2005;37:1217–1223. - PubMed
- Nannya Y, Taura K, Kurokawa M, Chiba S, Ogawa S. Evaluation of genome-wide power of genetic association studies based on empirical data from the HapMap project. Hum Mol Genet. 2007;16:3494–3505. - PubMed
- Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet. 2007;39:906–913. - PubMed
Publication types
MeSH terms
Grants and funding
- R01 AG026564/AG/NIA NIH HHS/United States
- R21 AG027110/AG/NIA NIH HHS/United States
- P50 AR055081/AR/NIAMS NIH HHS/United States
- R21 AA015973/AA/NIAAA NIH HHS/United States
- R01 AR050496/AR/NIAMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Research Materials