Targeted screening of cis-regulatory variation in human haplotypes - PubMed (original) (raw)
doi: 10.1101/gr.084798.108. Epub 2008 Oct 29.
Bing Ge, Elin Grundberg, Rose Hoberman, Kevin C L Lam, Vonda Koka, Joana Dias, Scott Gurd, Nicolas W Martin, Hans Mallmin, Olof Nilsson, Eef Harmsen, Ken Dewar, Tony Kwan, Tomi Pastinen
Affiliations
- PMID: 18971308
- PMCID: PMC2612965
- DOI: 10.1101/gr.084798.108
Targeted screening of cis-regulatory variation in human haplotypes
Dominique J Verlaan et al. Genome Res. 2009 Jan.
Abstract
Regulatory cis-acting variants account for a large proportion of gene expression variability in populations. Cis-acting differences can be specifically measured by comparing relative levels of allelic transcripts within a sample. Allelic expression (AE) mapping for cis-regulatory variant discovery has been hindered by the requirements of having informative or heterozygous single nucleotide polymorphisms (SNPs) within genes in order to assign the allelic origin of each transcript. In this study we have developed an approach to systematically screen for heritable cis-variants in common human haplotypes across >1,000 genes. In order to achieve the highest level of information per haplotype studied, we carried out allelic expression measurements by using both intronic and exonic SNPs in primary transcripts. We used a novel RNA pooling strategy in immortalized lymphoblastoid cell lines (LCLs) and primary human osteoblast cell lines (HObs) to allow for high-throughput AE. Screening hits from RNA pools were further validated by performing allelic expression mapping in individual samples. Our results indicate that >10% of expressed genes in human LCLs show genotype-linked AE. In addition, we have validated cis-acting variants in over 20 genes linked with common disease susceptibility in recent genome-wide studies. More generally, our results indicate that RNA pooling coupled with AE read-out by second generation sequencing or by other methods provides a high-throughput tool for cataloging the impact of common noncoding variants in the human genome.
Figures
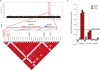
Figure 1.
(A) Hypothetical Manhattan plot for a disease GWAS, demonstrating a strong association to SNPs on chromosome 14, highlighted in red block. (B) Genomic region demonstrating an LD block spanning the GWAS SNPs. Marker SNPs (m1SNP, m2SNP, m3SNP) in genes (A and B), found in strong LD with the GWAS SNP, are used to assay for AE (rSNP, regulatory SNP). (C) Normalized allele ratio (log10) graph demonstrating different scenarios. Strong allelic expression is found for Gene A using m1SNP that tests for the same haplotypes found for the GWAS SNP. Marker m2SNP shows borderline AE for the same gene (Gene A) and tests for a different pattern of haplotypes. No AE is found for Gene B using m3SNP as the gDNA, and cDNA ratios are not significantly different from each other.
Figure 2.
Allelic imbalance association mapping results for GWAS genes. Vertical red lines correspond to –log10(_P_-value) of the AE assay for each SNP tested. The arrow points to the GWAS SNPs, indicated by the vertical blue lines. (A) GSDMB (asthma); (B) ORMDL3 (asthma); (C) IL23R (Crohn’s disease); (D) SORT1 (Dyslipidemia); (E) INSIG2 (Obesity); (F) TRAF1 (Rheumatoid arthritis).
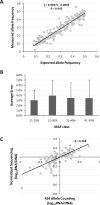
Figure 3.
(A) Comparison of expected and observed allele count (n = 278) in DNA pools derived from 55 CEU LCLs. (B) Average error with standard deviation (error bars) of allele frequencies derived from 454 counting as compared with known frequencies in the DNA pool. (C) Correlation of estimated fold difference (log10) in allele frequencies between cDNA and gDNA pools based on normalized sequencing (_y_-axis) and 454 allele counting (_x_-axis).
Similar articles
- Mapping common regulatory variants to human haplotypes.
Pastinen T, Ge B, Gurd S, Gaudin T, Dore C, Lemire M, Lepage P, Harmsen E, Hudson TJ. Pastinen T, et al. Hum Mol Genet. 2005 Dec 15;14(24):3963-71. doi: 10.1093/hmg/ddi420. Epub 2005 Nov 21. Hum Mol Genet. 2005. PMID: 16301213 - A survey of genetic and epigenetic variation affecting human gene expression.
Pastinen T, Sladek R, Gurd S, Sammak A, Ge B, Lepage P, Lavergne K, Villeneuve A, Gaudin T, Brändström H, Beck A, Verner A, Kingsley J, Harmsen E, Labuda D, Morgan K, Vohl MC, Naumova AK, Sinnett D, Hudson TJ. Pastinen T, et al. Physiol Genomics. 2004 Jan 15;16(2):184-93. doi: 10.1152/physiolgenomics.00163.2003. Physiol Genomics. 2004. PMID: 14583597 - Allele-specific transcript quantification detects haplotypic variation in the levels of the SDF-1 transcripts.
Kimura R, Nishioka T, Soemantri A, Ishida T. Kimura R, et al. Hum Mol Genet. 2005 Jun 15;14(12):1579-85. doi: 10.1093/hmg/ddi166. Epub 2005 Apr 20. Hum Mol Genet. 2005. PMID: 15843397 - Cis-acting regulatory variation in the human genome.
Pastinen T, Hudson TJ. Pastinen T, et al. Science. 2004 Oct 22;306(5696):647-50. doi: 10.1126/science.1101659. Science. 2004. PMID: 15499010 Review. - Influence of human genome polymorphism on gene expression.
Pastinen T, Ge B, Hudson TJ. Pastinen T, et al. Hum Mol Genet. 2006 Apr 15;15 Spec No 1:R9-16. doi: 10.1093/hmg/ddl044. Hum Mol Genet. 2006. PMID: 16651375 Review.
Cited by
- Gene-gene interaction and functional impact of polymorphisms on innate immune genes in controlling Plasmodium falciparum blood infection level.
Basu M, Das T, Ghosh A, Majumder S, Maji AK, Kanjilal SD, Mukhopadhyay I, Roychowdhury S, Banerjee S, Sengupta S. Basu M, et al. PLoS One. 2012;7(10):e46441. doi: 10.1371/journal.pone.0046441. Epub 2012 Oct 12. PLoS One. 2012. PMID: 23071570 Free PMC article. - Characterizing the genetic basis of transcriptome diversity through RNA-sequencing of 922 individuals.
Battle A, Mostafavi S, Zhu X, Potash JB, Weissman MM, McCormick C, Haudenschild CD, Beckman KB, Shi J, Mei R, Urban AE, Montgomery SB, Levinson DF, Koller D. Battle A, et al. Genome Res. 2014 Jan;24(1):14-24. doi: 10.1101/gr.155192.113. Epub 2013 Oct 3. Genome Res. 2014. PMID: 24092820 Free PMC article. - Epigenetic modifications are associated with inter-species gene expression variation in primates.
Zhou X, Cain CE, Myrthil M, Lewellen N, Michelini K, Davenport ER, Stephens M, Pritchard JK, Gilad Y. Zhou X, et al. Genome Biol. 2014;15(12):547. doi: 10.1186/s13059-014-0547-3. Genome Biol. 2014. PMID: 25468404 Free PMC article. - Signatures of Evolutionary Adaptation in Quantitative Trait Loci Influencing Trace Element Homeostasis in Liver.
Engelken J, Espadas G, Mancuso FM, Bonet N, Scherr AL, Jímenez-Álvarez V, Codina-Solà M, Medina-Stacey D, Spataro N, Stoneking M, Calafell F, Sabidó E, Bosch E. Engelken J, et al. Mol Biol Evol. 2016 Mar;33(3):738-54. doi: 10.1093/molbev/msv267. Epub 2015 Nov 17. Mol Biol Evol. 2016. PMID: 26582562 Free PMC article. - Integrated genetic and epigenetic analysis identifies haplotype-specific methylation in the FTO type 2 diabetes and obesity susceptibility locus.
Bell CG, Finer S, Lindgren CM, Wilson GA, Rakyan VK, Teschendorff AE, Akan P, Stupka E, Down TA, Prokopenko I, Morison IM, Mill J, Pidsley R; International Type 2 Diabetes 1q Consortium; Deloukas P, Frayling TM, Hattersley AT, McCarthy MI, Beck S, Hitman GA. Bell CG, et al. PLoS One. 2010 Nov 18;5(11):e14040. doi: 10.1371/journal.pone.0014040. PLoS One. 2010. PMID: 21124985 Free PMC article.
References
- Bouatia-Naji N., Rocheleau G., Van Lommel L., Lemaire K., Schuit F., Cavalcanti-Proenca C., Marchand M., Hartikainen A.L., Sovio U., De Graeve F., et al. A polymorphism within the G6PC2 gene is associated with fasting plasma glucose levels. Science. 2008;320:1085–1088. - PubMed
- Bray N.J., Buckland P.R., Owen M.J., O'Donovan M.C. Cis-acting variation in the expression of a high proportion of genes in human brain. Hum. Genet. 2003;113:149–153. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials