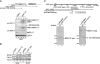The role of the putative 3' end processing endonuclease Ysh1p in mRNA and snoRNA synthesis - PubMed (original) (raw)
The role of the putative 3' end processing endonuclease Ysh1p in mRNA and snoRNA synthesis
Monika Garas et al. RNA. 2008 Dec.
Abstract
Pre-mRNA 3' end formation is tightly linked to upstream and downstream events of eukaryotic mRNA synthesis. The two-step reaction involves endonucleolytic cleavage of the primary transcript followed by poly(A) addition to the upstream cleavage product. To further characterize the putative 3' end processing endonuclease Ysh1p/Brr5p, we isolated and analyzed a number of new temperature- and cold-sensitive mutant alleles. We show that Ysh1p plays a crucial role in 3' end formation and in RNA polymerase II (RNAP II) transcription termination on mRNA genes. In addition, we observed a range of additional functional deficiencies in ysh1 mutant strains, which were partially allele-specific. Interestingly, snoRNA 3' end formation and RNAP II termination were defective on specific snoRNAs in the cold-sensitive ysh1-12 strain. Moreover, we observed the accumulation of several mRNAs including the NRD1 transcript in this mutant. We provide evidence that NRD1 autoregulation is associated with endonucleolytic cleavage and that this process may involve Ysh1p. In addition, the ysh1-12 strain displayed defects in RNA splicing indicating that a functional link may exist between intron removal and 3' end formation in yeast. These observations suggest that Ysh1p has multiple roles in RNA synthesis and processing.
Figures
FIGURE 1.
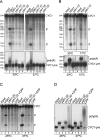
Isolation of conditional ysh1 mutants. (A) The conserved β-lactamase signature H68X69H70X71D72H73 is essential for cell viability. Plasmid shuffling was used to test the requirement of H68, H70, and D72 residues of the β-lactamase consensus motif for cell viability. The LM109 strain with a disrupted chromosomal YSH1 gene and carrying _YSH1_-URA3 plasmid was transformed with the _ADE2_-plasmid bearing either the wild-type YSH1 gene or its mutant versions H68A, H70A, and D72A, followed by counterselection on 5′-FOA plates. (B) Tenfold serial dilutions of cultures of wild-type and ysh1 mutant strains were spotted onto YPAD medium and incubated for 3 d at 23°–37°C or 5 d at 15°C. (C) Schematic representation of ysh1 mutant sequences underlying the respective temperature- or cold-sensitive phenotypes. (Shaded boxes) The β-lactamase, β-CASP, and C-terminal domains and the conserved H68F69H70L71D72H73 signature are marked approximately.
FIGURE 2.
Ysh1p is required for cleavage and polyadenylaion of pre-mRNA in vitro. (A, upper panel) In vitro cleavage and (lower panel) polyadenylation assays with protein extracts prepared from wild-type and ysh1 temperature-sensitive strains as indicated. Input shows a control where no protein was added. Positions of substrate RNAs, 5′ and 3′ end cleavage products and polyadenylation products bands are shown. HpaII-digested pBR322 fragments were 5′ end labeled and served as markers. Internally [32P]-labeled substrate RNAs, CYC1 for the cleavage assay and CYC1-pre for specific polyadenylation, were used. Reactions were performed either at 30°C (lanes 1–6) or at 37°C (lanes 7–12). Extracts were preincubated at restrictive temperatures for 10 min prior to assaying. (B) As in A, except reactions were performed either at the permissive temperature 30°C (lanes 1–3) or at the non-permissive temperatures 17°C (lanes 4–6) for cleavage, and at 15°C for polyadenylation. (C,D) Reconstitution of specific cleavage and polyadenylation activities of the ysh1-32 extract in vitro. (C) Cleavage and (D) polyadenylation assays were performed essentially as in A, with protein extracts prepared from wild-type and the ysh1-32 strain as indicated. The ysh1-32 extract was combined with purified CPF to restore the specific 3′end processing activity. Twice more CPF was used for the reconstitution of polyadenylation than for the reconstitution of cleavage.
FIGURE 3.
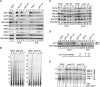
Ysh1p is required for normal mRNA accumulation and poly(A) site selection. (A) Northern blot analysis of mRNA steady-state levels in ysh1 strains. Wild-type, _ysh1_-32, and rna15-1 strains were grown in YPD at 25°C and shifted for 1, 3, and 6 h to 37°C. The cold-sensitive strain ysh1-12 was grown in YPD at 30°C and shifted for 1, 3 and 6 h to 15°C. Total RNA was extracted from wild-type and mutant ysh1 cells and separated on formaldehyde/1.2% agarose gels. Filters were developed with either random-primed labeled DNA probes or end-labeled oligonucleotides directed against RNA species indicated on the left. 18S rRNA and SCR1 served as loading controls. (B) Poly(A) tails shorten in the ysh1-32 strain after shift to restrictive temperature. 3′ End labeling of poly(A) tails with total RNA extracted from (left panel) wild-type and ysh1-32 or (right panel) wild-type and ysh1-12 mutant strains after growth at permissive temperatures (23°C or 30°C, respectively), and after shift for 1, 3, and 6 h to respective restrictive temperatures (37°C or 15°C). Poly(A) tail length (in nucleotides) is indicated on the left. (C) Western blot analysis of wild-type, ysh1-32, and ysh1-12 mutant extracts prepared from cells grown analogously to A. Equal amounts of total protein were loaded in each lane. Blots were probed with antibodies directed against the proteins indicated on the left. (D) Northern blot analysis of total RNAs extracted from wild-type and mutant ysh1 cells. The ysh1-13, ysh1-15, ysh1-32, and rna15-1 strains were grown in YPD at 23°C and shifted for 1, 3, and 6 h to 37°C. ysh1-12 was grown at 30°C and shifted for 1, 3, and 6 h to 15°C. The analysis was performed essentially as in A. 18S rRNA served as a loading control. The positions of ACT1 poly(A) sites I–V are labeled. The scheme below the panel represents the relative positions of the different ACT1 poly(A) sites. (E) Analysis of ACT1 poly(A) site usage in wild-type, ysh1-13, and ysh1-15 cells. Total RNAs extracted from strains grown as described in E were treated with RNase H and oligonucleotides _ACT1_-RNase H and oligo(dT), and analyzed by polyacrylamide Northern blotting with a probe specific for the 3′ end of ACT1 mRNA. Positions of the poly(A) sites I–V are indicated on the right. RNA levels were quantified by PhosphorImager scanning (Molecular Dynamics). The ratios of poly(A) site I versus site V usage for each lane are indicated at the bottom.
FIGURE 4.
Mutations in YSH1 impair correct RNAP II transcription termination on mRNA genes in vivo. (A) Slot hybridizations of run-on transcripts obtained after transcriptional run-on analysis (TRO) (Birse et al. 1998). The scheme represents the p_GAL-CYC1_ gene construct that was used for TRO analysis. The order of M13 probes spanning the CYC1 gene (P1–P6) and poly(A) site (position 506) are depicted. Wild-type and ysh1-12 cells transformed with p_UGCYC1_ plasmid were grown in synthetic medium lacking uracil and containing 2% galactose under permissive growth conditions (30°C) and after shift for 1 h to restrictive temperature (15°C). P1–P6 represent probes complementary to CYC1 transcripts as indicated on the scheme. Empty M13 was used as a background hybridization control. Hybridization of RNAP III transcripts to the tRNA probe is shown. (B) Quantitative analysis of transcriptional run-on profiles for the ysh1-12 mutant. Values obtained by PhosphorImager scanning (Molecular Dynamics) were corrected by subtraction of the M13 background signal and normalized to the value of P1, which was fixed at 100%. Results shown in the diagram represent the average value of three independent experiments. (C) Quantitative analysis of transcriptional run-on profiles for the _ysh1_-32 strain. Values were obtained as described in B. (D,E) Read-through mRNA transcripts accumulate in the ysh1-12 strain in vivo. Northern blot analysis of total RNA extracted from wild-type and ysh1-12 cells (lanes 1–3,7–9) grown in YPD medium at permissive temperature 30°C and (lanes 4–6,10–12) followed by a shift for 1, 3 or 6 h to 15°C. RNAs were resolved as described in Figure 3A. The filter was developed with random-primed labeled probe directed against (D) YSH1 or (E) ADH1, then subsequently washed and probed for the RNA of the downstream gene (DBP9 or YOL087C, respectively), as indicated. (*) Unidentified intermediate-sized transcripts. Probing to the RNAP III transcript SCR1 served as a loading control. (Bottom) Schematic representation of the (D) YSH1-DBP9 and the (E) _ADH1_-YOL087C gene loci.
FIGURE 5.
Mutations in ysh1-12 impair snoRNA 3′ end formation and transcription termination. (A) 3′ end extended snoRNA transcripts are produced in the ysh1-12 mutant strain. Primer extension (PE) analysis of extended transcripts of several snoRNAs in the wild-type and ysh1-12 strains was carried out with radioactively labeled oligonucleotides complementary to sequences located downstream from the mature 3′ ends of the indicated snoRNAs. An oligonucleotide complementary to the mature U3 snoRNA was used as control. (B) Transcription termination on the SNR3 snoRNA gene is impaired in _ysh1_-12. Slot hybridizations of run-on transcripts were obtained after TRO. (Top) The scheme represents the SNR3-YJR129c genomic locus and location of the TRO probes A and B. Note that SNR3 and YJR129c are transcribed from opposite strands of the DNA, as indicated by the arrows. (Lanes 1,2) Wild-type and ysh1-12 cells were grown in YPD medium containing 2% glucose under permissive growth conditions (30°C) and (lanes 3,4) after the shift to restrictive temperature (15°C) for 1 h. M13 slots are single-stranded phagemids with no insert and were used as a background hybridization control. The ACT1 probe served as a positive control for hybridization. (C) Quantitative analysis of transcriptional run-on profiles for the ysh1-12 mutant. Values obtained by PhosphorImager scanning (Molecular Dynamics) were corrected by subtraction of the M13 background signal and normalized to the value of probe A, which was set at 100%. Results shown in the diagram represent the average value of three independent experiments. (D) Western blot analysis of isogenic wild-type (FY23) and brr5-td degron strain extracts prepared from cells grown at 25°C and shifted for 1, 2, or 3 h to 37°C. Equal amounts of total protein were loaded in each lane. The blot was probed with antibody directed against Ysh1p and subsequently re-probed with antibodies directed against other CPF subunits, as indicated. (E) Primer extension analysis of extended transcripts of several snoRNAs in the wild-type FY23, brr5-td (both shifted for 2 h to 37°C), and ysh1-12 (shifted for 6 h to 15°C) strains was carried out with radioactively labeled oligonucleotides complementary to sequences located downstream from the mature 3′ ends of the indicated snoRNAs. (Lanes 1,3) Levels of the RNAP III transcript U6 served as a loading control and showed that lower amounts of total RNA extracted from the FY23 and brr5-td strains were used in this experiment.
FIGURE 6.
Involvement of Ysh1p in the autoregulation of NRD1 mRNA levels. (A) Increased levels of NRD1 mRNA accumulate in the ysh1-12 mutant strain. Northern blot analysis of NRD1 mRNA in wild-type and ysh1-12 strains was performed as in Figure 3A, with probes directed against the 5′ end of NRD1 mRNA, or against MRPL17, as indicated on the left. (Top) The scheme represents the _NRD1_-MRPL17 gene loci; transcripts originating from the NRD1 transcription start are indicated. Hybridization to the RNAP III transcript SCR1 served as a loading control. (B) The amount of Nrd1 protein increases in ysh1-12 cells. Western blot analysis of wild-type and mutant extracts prepared from cells grown in YPD at 25°C and shifted for 3 h to restrictive temperatures (15°C or 37°C, respectively). Equal amounts of total protein were loaded in each lane. Blots were probed with antibodies directed against the proteins indicated on the left. Act1p levels served as control for equal loading. (C) NRD1 premature termination occurs via an endonucleolytic cleavage of the NRD1 pre-mRNA. (Upper part) Schematic representation of the NRD1 gene locus. Positions of the oligonucleotides U (+300 nt) and D (+1300 nt; relative to the ORF start) used in the primer extension analyses, the full-length and prematurely terminated NRD1 mRNA transcripts, and of the putative 3′ end cutoff product are shown. (Below, right panel) Primer extension analysis of total RNA extracted from wild-type (W303) and _rat1_-1, xrn1Δ strains (grown for 2 h at 37°C), carried out with radioactively labeled oligonucleotide D complementary to sequences located downstream from the predicted premature termination region. (Left panel) Oligonucleotide U complementary to the 5′ end of NRD1 transcript was used as an internal control.
FIGURE 7.
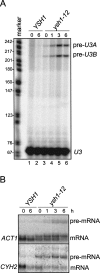
The ysh1-12 mutant is defective in splicing. (A) Primer extension analysis of spliced and unspliced U3 transcripts in wild-type and ysh1-12 mutant strains grown in YPD for up to 6 h at 15°C. Analysis was carried out with a radioactively labeled oligonucleotide complementary to sequences located in U3 exon 2. HpaII-digested pBR322 fragments were 5′ end labeled and served as markers. (Right) Bands corresponding to pre-U3A, pre-U3B, and mature U3 snoRNA are indicated. (B) Northern blot analysis of total RNA extracted from wild-type and mutant ysh1-12 cells, performed as in Figure 3A. Filters were developed with random-primed labeled probes directed against RNA transcripts as indicated on the left.
Similar articles
- Pti1p and Ref2p found in association with the mRNA 3' end formation complex direct snoRNA maturation.
Dheur S, Vo le TA, Voisinet-Hakil F, Minet M, Schmitter JM, Lacroute F, Wyers F, Minvielle-Sebastia L. Dheur S, et al. EMBO J. 2003 Jun 2;22(11):2831-40. doi: 10.1093/emboj/cdg253. EMBO J. 2003. PMID: 12773397 Free PMC article. - Yeast snoRNA accumulation relies on a cleavage-dependent/polyadenylation-independent 3'-processing apparatus.
Fatica A, Morlando M, Bozzoni I. Fatica A, et al. EMBO J. 2000 Nov 15;19(22):6218-29. doi: 10.1093/emboj/19.22.6218. EMBO J. 2000. PMID: 11080167 Free PMC article. - Polyadenylation linked to transcription termination directs the processing of snoRNA precursors in yeast.
Grzechnik P, Kufel J. Grzechnik P, et al. Mol Cell. 2008 Oct 24;32(2):247-58. doi: 10.1016/j.molcel.2008.10.003. Mol Cell. 2008. PMID: 18951092 Free PMC article. - A history of poly A sequences: from formation to factors to function.
Edmonds M. Edmonds M. Prog Nucleic Acid Res Mol Biol. 2002;71:285-389. doi: 10.1016/s0079-6603(02)71046-5. Prog Nucleic Acid Res Mol Biol. 2002. PMID: 12102557 Review. - Polyadenylation: a tail of two complexes.
Proudfoot N, O'Sullivan J. Proudfoot N, et al. Curr Biol. 2002 Dec 23;12(24):R855-7. doi: 10.1016/s0960-9822(02)01353-2. Curr Biol. 2002. PMID: 12498707 Review.
Cited by
- Transcription termination by nuclear RNA polymerases.
Richard P, Manley JL. Richard P, et al. Genes Dev. 2009 Jun 1;23(11):1247-69. doi: 10.1101/gad.1792809. Genes Dev. 2009. PMID: 19487567 Free PMC article. - Advances in the mechanism of small nucleolar RNA and its role in DNA damage response.
Shen LP, Zhang WC, Deng JR, Qi ZH, Lin ZW, Wang ZD. Shen LP, et al. Mil Med Res. 2024 Aug 8;11(1):53. doi: 10.1186/s40779-024-00553-4. Mil Med Res. 2024. PMID: 39118131 Free PMC article. Review. - Transcription Dynamics in Living Cells.
Lenstra TL, Rodriguez J, Chen H, Larson DR. Lenstra TL, et al. Annu Rev Biophys. 2016 Jul 5;45:25-47. doi: 10.1146/annurev-biophys-062215-010838. Epub 2016 Apr 27. Annu Rev Biophys. 2016. PMID: 27145880 Free PMC article. Review. - Pcf11 orchestrates transcription termination pathways in yeast.
Grzechnik P, Gdula MR, Proudfoot NJ. Grzechnik P, et al. Genes Dev. 2015 Apr 15;29(8):849-61. doi: 10.1101/gad.251470.114. Epub 2015 Apr 15. Genes Dev. 2015. PMID: 25877920 Free PMC article. - Genetic and pharmacological evidence for kinetic competition between alternative poly(A) sites in yeast.
Turner RE, Harrison PF, Swaminathan A, Kraupner-Taylor CA, Goldie BJ, See M, Peterson AL, Schittenhelm RB, Powell DR, Creek DJ, Dichtl B, Beilharz TH. Turner RE, et al. Elife. 2021 Jul 7;10:e65331. doi: 10.7554/eLife.65331. Elife. 2021. PMID: 34232857 Free PMC article.
References
- Aravind L. An evolutionary classification of the metallo-β-lactamase fold proteins. In Silico Biol. 1999;1:69–91. - PubMed
- Arigo J.T., Carroll K.L., Ames J.M., Corden J.L. Regulation of yeast NRD1 expression by premature transcription termination. Mol. Cell. 2006;21:641–651. - PubMed
- Bentley D.L. Rules of engagement: Co-transcriptional recruitment of pre-mRNA processing factors. Curr. Opin. Cell Biol. 2005;17:251–256. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases