A formal ontology of subcellular neuroanatomy - PubMed (original) (raw)
A formal ontology of subcellular neuroanatomy
Stephen D Larson et al. Front Neuroinform. 2007.
Abstract
The complexity of the nervous system requires high-resolution microscopy to resolve the detailed 3D structure of nerve cells and supracellular domains. The analysis of such imaging data to extract cellular surfaces and cell components often requires the combination of expert human knowledge with carefully engineered software tools. In an effort to make better tools to assist humans in this endeavor, create a more accessible and permanent record of their data, and to aid the process of constructing complex and detailed computational models, we have created a core of formalized knowledge about the structure of the nervous system and have integrated that core into several software applications. In this paper, we describe the structure and content of a formal ontology whose scope is the subcellular anatomy of the nervous system (SAO), covering nerve cells, their parts, and interactions between these parts. Many applications of this ontology to image annotation, content-based retrieval of structural data, and integration of shared data across scales and researchers are also described.
Keywords: data integration; electron microscopy; neuroanatomy; neuronal cell types; subcellular anatomy.
Figures
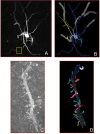
Figure 1
Multiple representations of the same medium spiny neuron taken from the CCDB. In (A), a light-level fill of the neuron. The yellow box shows the portion of the dendritic branch shown in (C). In (B), the Neurolucida segmentation of that neuron. In (C), the EM image of the portion of the dendrite featured in (A). In (D), the 3D reconstruction of the dendrite from (C) after segmentation.
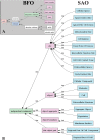
Figure 2
High level class structure of the SAO. The BFO entities are shown in (A) and in the green and pink boxes in (B). Spatial regions, subclasses of occurrents, and subclasses of realizable entity have been omitted because the SAO does not currently use them. SAO classes that are under the BFO hierarchy are shown in blue in (B).
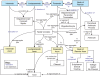
Figure 3
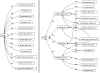
Diagram of a Node of Ranvier instance description in the SAO. The boxes indicate instances of classes that are related to one another as a description of a particular instance of a Node of Ranvier. The blue text indicates relationships that are enforced between classes through the use of OWL restrictions, while the black text indicates relationships defined for this instance alone.
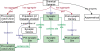
Figure 4
Diagram of a chemical synapse instance description in SAO. Sites are indicated by green backgrounds. The boxes indicate Instances of classes that are related to one another as a description of a particular instance of a chemical synapse.
Figure 5
Inferred hierarchies using OWL. On the left, a subset of the hierarchy under the Neuron class prior to inference. On the right, the automatic reclassification of that subset under four user-defined groupings, GlutamatergicNeuron, GABAergicNeuron, SpinyCell, GranuleCell, based on the properties of the cells alone.
References
- Bota M., Dong H. W., Swanson L. W. (2005). Brain architecture management system. Neuroinformatics 3, 15–48 - PubMed
- Bowden D. M., Dubach M. F. (2003). NeuroNames 2002. Neuroinformatics 1, 43–59 - PubMed
- Chen L., Martone M. E., Gupta A., Fong L., Wong-Barnum M. (2006). OntoQuest: exploring ontological data made easy. Abstract, Very Large Databases Conference1183–1186
Grants and funding
- R01 NS058296/NS/NINDS NIH HHS/United States
- P41 LM007885/LM/NLM NIH HHS/United States
- U24 RR021760/RR/NCRR NIH HHS/United States
- P41 RR004050/RR/NCRR NIH HHS/United States
- R01 DA016602/DA/NIDA NIH HHS/United States
- P41 RR008605/RR/NCRR NIH HHS/United States
LinkOut - more resources
Full Text Sources