Intraspecific evolution of the intercellular signaling network underlying a robust developmental system - PubMed (original) (raw)
Intraspecific evolution of the intercellular signaling network underlying a robust developmental system
Josselin Milloz et al. Genes Dev. 2008.
Abstract
Many biological systems produce an invariant output when faced with stochastic or environmental variation. This robustness of system output to variation affecting the underlying process may allow for "cryptic" genetic evolution within the system without change in output. We studied variation of cell fate patterning of Caenorhabditis elegans vulva precursors, a developmental system that relies on a simple intercellular signaling network and yields an invariant output of cell fates and lineages among C. elegans wild isolates. We first investigated the system's genetic variation in C. elegans by means of genetic tools and cell ablation to break down its buffering mechanisms. We uncovered distinct architectures of quantitative variation along the Ras signaling cascade, including compensatory variation, and differences in cell sensitivity to induction along the anteroposterior axis. In the unperturbed system, we further found variation between isolates in spatio-temporal dynamics of Ras pathway activity, which can explain the phenotypic differences revealed upon perturbation. Finally, the variation mostly affects the signaling pathways in a tissue-specific manner. We thus demonstrate and characterize microevolution of a developmental signaling network. In addition, our results suggest that the vulva genetic screens would have yielded a different mutation spectrum, especially for Wnt pathway mutations, had they been performed in another C. elegans genetic background.
Figures
Figure 1.
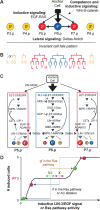
Uncovering cryptic genetic variation in C. elegans vulva development. (A) C. elegans vulva development. The invariant cell fate pattern (3°3°2°1°2°3°) is established through the interplay of three cell signaling pathways: Egf–Egfr–Ras, Delta–Notch, and Wnt–β-catenin. (B) Vulval precursor cell lineage. Letters indicate the division plane: (T) transverse (left–right), (L) longitudinal (anteroposterior), (U) undivided. The primary lineage is characterized by division of all four grandaughters in a transverse orientation. The secondary lineage (“
L
LTU” or “UTL
L
”) is characterized by an absence of division of one granddaughter and adhesion to the cuticle (underline). The tertiary lineage is a nonvulval fate, with a single division (“S”, fusion with the epidermal syncytium hyp7). The 3°3°2°1°2°3° pattern and Pn.p cell lineage are invariant in all wild isolates (except for P3.p division frequency) (Delattre and Félix 2001). (C) Vulva signaling network showing the different Ras pathway components and the interplay between Ras and Notch pathways (see main text for description). (D) Schematic representation of invasive approaches to uncover cryptic genetic variation in the system. The graph shows a theoretical curve of the number of induced Pn.p cells (1° or 2° fate) as a function of EGF dose or Ras pathway activity; this number may range from 0 (no anchor cell) to 6 (egf overexpression). The plateau at the wild-type phenotype of three induced cells materializes robustness to some variation in the EGF signal or Ras pathway activity (Braendle et al. 2008; Félix and Wagner 2008). While N2, the reference C. elegans wild isolate, is located on the plateau under standard conditions, a second isolate (X) could be located at a different position on the plateau (stasis in the final fate pattern). Applying the same perturbation to both isolates will schematically shift their respective positions along the _X_-axis, thus uncovering an initially silent difference. As perturbations, we applied mutations in the Ras pathway (Figs. 2, 3) and anchor cell ablations (Fig. 4). (gf) Gain of function; (rf) reduction of function.
Figure 2.
Expressivity of vulval induction defects of Ras, Notch, and Wnt pathway mutations in wild C. elegans backgrounds. We initially chose one mutation per pathway, with an effect on vulval development and no strong lethal effect, and introgressed it into seven wild genetic backgrounds of C. elegans. (A) Ras pathway: let-23(sy1rf)/egfr. The expressivity of this mutation varies significantly among wild C. elegans backgrounds (ANOVA, main effect isolate F6,455 = 21.94, P < 10−4, n = 49–70). (B) Notch pathway: sup-17(n316rf)/adam10 (ANOVA, main effect isolate: F6,441 = 5.34, P < 10−4, n = 41–91). (C) Wnt pathway: bar-1(ga80lf)/ β-catenin (ANOVA, main effect isolate: F6,1657 = 20.38, P < 10−4, n = 187–347). (rf) Reduction of function; (lf) loss of function. (D) Wild-type vulva at the L4 stage under Nomarski optics. The three induced Pn.p cells give rise to 22 progeny. (E,F) Representative mutant phenotypes of animals carrying the let-23(sy1rf)/egfr mutation in the N2 (E) and AB1 (F) backgrounds. Half a Pn.p cell is induced (the posterior daughter of P6.p) in E and 2.5 Pn.p cells in F. Underline indicates vulval tissue. For each mutation, we performed ANOVA tests for the effect of Isolate (the wild genetic background) and of Replicate nested in Isolate. As the effect of Isolate was statistically significant, we next performed post-hoc Tukey’s HSD test on Isolate to determine groups of statistical significance (Supplemental Table S4): For each mutation, the mean number of induced cells is significantly different between isolates labeled with different letters (P < 0.05) (Supplemental Table S4). Error bars indicate standard error of the mean (SE) over replicates.
Figure 3.
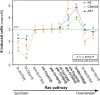
Expressivity of vulval induction defects caused by mutations at different steps along the EGFR-Ras pathway in the N2, CB4856, and AB1 wild genetic backgrounds. The graph shows the number of induced cells (_Y_-axis, mean and standard error of replicates) for different mutations along the Ras pathway (_X_-axis, upstream to downstream parts of the pathway from left to right) in three wild genetic backgrounds (colors). Bold lettering indicates mutations that hyperactivate the pathway, while nonbold lettering refers to mutations that underactivate the pathway. See Figure 1C for a schematic drawing of the Ras pathway and its regulators. (rf) Reduction of function (hypomorph); (lf) loss of function (putative null); (gf) gain of function. The lin-3 and let-23 alleles are tissue specific and strong hypomorphs for the vulva, with no effect on lethality; the other reduction-of-function mutations in positive regulators of the pathway (sem-5, lin-45, and mpk-1 alleles) in addition result in L1 lethality (see Fig. 6) and are milder hypomorphs for the vulva. The other alleles (gain of function in Ras, or loss of function of negative regulators) result in gain of activity in the pathway (more than three cells induced). For each mutation, the mean number of induced cells is significantly different between isolates labeled with different letters (Tukey’s HSD test for multiple isolate comparisons and ANOVA main effect Isolate for two isolate comparisons; P < 0.05, n = 49–210) (see Fig. 2 legend and Supplemental Table S4 for more explanations).
Figure 4.
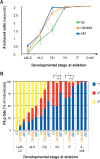
Effect of anchor cell ablation in the N2, CB4856, and AB1 wild isolates. (A) Effect of anchor cell ablation on vulval induction in different C. elegans wild isolates over time of ablation. The anchor cell was ablated at different time points (_X_-axis; n ≥ 20 individuals per time point, per isolate) and the resulting vulval lineage followed in the early L4 stage. Distributions of the mean number of induced cells differ among isolates (ANOVA, effect isolate: F2,285 = 9.96, P < 10−4) (Supplemental Table S8). CB4856 and AB1 differ from N2 but not from one another (Tukey’s HSD post-hoc test on isolate, P < 0.05). (B) Effect of anchor cell ablation on the cell fate adopted by P6.p in the same experiment. For each time point, the bar shows the proportion of 1° (blue), 2° (red), and 3° (yellow) fates adopted by P6.p in different ablated individuals. χ2 test for the proportion of 1° versus 2° cell fates (almost no 3° cell fate at these time points), (*) P < 0.05. In this test, AB1 is significantly different from both N2 and CB4856. Developmental stages: (Let. L2) lethargic L2; (eL3) early L3; (DU) Dorsal Uterine precursor cells dividing or divided once; (VU) Ventral Uterine precursor cells dividing or divided once; (3°) 3° cells have divided; (two-cell stage) all Pn.p cells have divided once. Cell lineages are shown in Supplemental Tables S5–S7.
Figure 5.
Quantification of downstream transcriptional reporters of the Ras and Notch pathways in the N2, CB4856, and AB1 wild isolates. (A) Micrographs showing the expression pattern of the Ras pathway reporter egl-17_∷_CFP and the Notch pathway reporter lip-1_∷_YFP at the early L3 stage in the same worm. (B) Summary diagram of the expression patterns of egl-17_∷_CFP (blue) and lip-1_∷_YFP (red). The egl-17∷CFP reporter is repressed over time in P5.p and P7.p, due to lateral Notch signaling. (C) Quantification of the Ras and Notch pathway reporters in three wild isolates. n = 10 individuals per time point, per isolate. Developmental stages are indicated on the left (see Fig. 4 for nomenclature). (f.u.) (Arbitrary) fluorescence units. We performed ANOVAs for each developmental stage to test for the effects of Isolate (the wild genetic background), Individual nested into Isolate [“Individual (isolate)”], Cell and Isolate by Cell interaction (“Isolate × Cell”) on CFP and on YFP signal intensity. Post-hoc Tukey’s HSD (P < 0.05) comparisons were then performed on the Isolate × Cell interaction to determine differences in signal intensity between isolates and cells (P5.p, P6.p, and P7.p). Within one graph, the mean signal intensity is significantly different between cells labeled with different letters. Error bars indicate standard error of the mean (SE) over replicates. For complete ANOVA results, see Supplemental Table S9.
Figure 6.
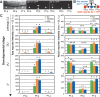
Variation in Wnt and Ras pathway mutation effects between tissues among C. elegans wild isolates. (A) Effect of bar-1(ga80lf)/β-catenin mutation on P12 transformation frequency and vulva defect expressivity in different wild genetic backgrounds (colors). The effect differs between genetic backgrounds for both P12 transformation (G-test, P < 10−4) and vulval defect expressivity (ANOVA, P < 10−4). (B) Effect of lin-45(n2018rf)/raf on L1 lethality, P12 transformation frequency and vulva defect expressivity. The effect in each of the three tissues varies between genetic backgrounds: P12 transformation (G-test, P < 10−4; combined probability for independent G-tests), L1 lethality (G-test, P < 10−4; combined probability for independent G-tests), and vulval defect expressivity (ANOVA, P < 10−4). (C) Effect of mpk-1(ku1rf)/mapk on L1 lethality and vulval defect expressivity. The effect differs between genetic backgrounds for both L1 lethality (G-test, P < 10−4; combined probability for independent G-tests) and vulval defect expressivity. (ANOVA, P < 10−4). (D) Effect of sem-5(n1779rf)/grb2 on L1 lethality and vulval defect expressivity. The effect differs between genetic backgrounds for both L1 lethality (G-test, P < 10−4; combined probability for independent G-tests) and vulval defect expressivity (ANOVA, P = 0.02). (P12 defect) Penetrance of P12 specification defects; (L1 lethality) penetrance of L1 lethality; (vulva) expressivity of vulval defect (vulval induction index). Phenotypic values for each tissue are scaled in each graph by ranking the values from mildest (bottom) to highest (top) penetrance or expressivity (with a minimum scale of 0.2 cell length for vulva data). Error bars indicate SE between replicates, when available. See Supplemental Table S12 for statistical analyses. Lines connect identical genetic backgrounds. Line crossing illustrates the lack of correlation between tissues.
Similar articles
- Role of pleiotropy in the evolution of a cryptic developmental variation in Caenorhabditis elegans.
Duveau F, Félix MA. Duveau F, et al. PLoS Biol. 2012 Jan;10(1):e1001230. doi: 10.1371/journal.pbio.1001230. Epub 2012 Jan 3. PLoS Biol. 2012. PMID: 22235190 Free PMC article. - [Genetic and environmental variations in an intercellular signaling network].
Félix MA. Félix MA. Med Sci (Paris). 2009 Aug-Sep;25(8-9):705-12. doi: 10.1051/medsci/2009258-9705. Med Sci (Paris). 2009. PMID: 19765384 Review. French. - Caenorhabditis elegans vulval cell fate patterning.
Félix MA. Félix MA. Phys Biol. 2012 Aug;9(4):045001. doi: 10.1088/1478-3975/9/4/045001. Epub 2012 Aug 7. Phys Biol. 2012. PMID: 22871570 - Cryptic quantitative evolution of the vulva intercellular signaling network in Caenorhabditis.
Félix MA. Félix MA. Curr Biol. 2007 Jan 23;17(2):103-14. doi: 10.1016/j.cub.2006.12.024. Curr Biol. 2007. PMID: 17240335 - Robustness and flexibility in nematode vulva development.
Félix MA, Barkoulas M. Félix MA, et al. Trends Genet. 2012 Apr;28(4):185-95. doi: 10.1016/j.tig.2012.01.002. Epub 2012 Feb 9. Trends Genet. 2012. PMID: 22325232 Review.
Cited by
- Chaperones, Canalization, and Evolution of Animal Forms.
Sato A. Sato A. Int J Mol Sci. 2018 Oct 4;19(10):3029. doi: 10.3390/ijms19103029. Int J Mol Sci. 2018. PMID: 30287767 Free PMC article. Review. - The future of evo-devo: model systems and evolutionary theory.
Sommer RJ. Sommer RJ. Nat Rev Genet. 2009 Jun;10(6):416-22. doi: 10.1038/nrg2567. Nat Rev Genet. 2009. PMID: 19369972 Review. - Cryptic genetic variation: evolution's hidden substrate.
Paaby AB, Rockman MV. Paaby AB, et al. Nat Rev Genet. 2014 Apr;15(4):247-58. doi: 10.1038/nrg3688. Epub 2014 Mar 11. Nat Rev Genet. 2014. PMID: 24614309 Free PMC article. Review. - Role of pleiotropy in the evolution of a cryptic developmental variation in Caenorhabditis elegans.
Duveau F, Félix MA. Duveau F, et al. PLoS Biol. 2012 Jan;10(1):e1001230. doi: 10.1371/journal.pbio.1001230. Epub 2012 Jan 3. PLoS Biol. 2012. PMID: 22235190 Free PMC article. - Does your gene need a background check? How genetic background impacts the analysis of mutations, genes, and evolution.
Chandler CH, Chari S, Dworkin I. Chandler CH, et al. Trends Genet. 2013 Jun;29(6):358-66. doi: 10.1016/j.tig.2013.01.009. Epub 2013 Feb 28. Trends Genet. 2013. PMID: 23453263 Free PMC article. Review.
References
- Abramoff M.D., Magelhaes P.J., Ram S.J. Image Processing with ImageJ. Biophotonics Int’l. 2004;11:36–42.
- Alon U., Surette M.G., Barkai N., Leibler S. Robustness in bacterial chemotaxis. Nature. 1999;397:168–171. - PubMed
- Barkai N., Leibler S. Robustness in simple biochemical networks. Nature. 1997;387:913–917. - PubMed
- Barrière A., Félix M.A. High local genetic diversity and low outcrossing rate in Caenorhabditis elegans natural populations. Curr. Biol. 2005;15:1176–1184. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources