A relation based measure of semantic similarity for Gene Ontology annotations - PubMed (original) (raw)
A relation based measure of semantic similarity for Gene Ontology annotations
Brendan Sheehan et al. BMC Bioinformatics. 2008.
Abstract
Background: Various measures of semantic similarity of terms in bio-ontologies such as the Gene Ontology (GO) have been used to compare gene products. Such measures of similarity have been used to annotate uncharacterized gene products and group gene products into functional groups. There are various ways to measure semantic similarity, either using the topological structure of the ontology, the instances (gene products) associated with terms or a mixture of both. We focus on an instance level definition of semantic similarity while using the information contained in the ontology, both in the graphical structure of the ontology and the semantics of relations between terms, to provide constraints on our instance level description.Semantic similarity of terms is extended to annotations by various approaches, either though aggregation operations such as min, max and average or through an extrapolative method. These approaches introduce assumptions about how semantic similarity of terms relates to the semantic similarity of annotations that do not necessarily reflect how terms relate to each other.
Results: We exploit the semantics of relations in the GO to construct an algorithm called SSA that provides the basis of a framework that naturally extends instance based methods of semantic similarity of terms, such as Resnik's measure, to describing annotations and not just terms. Our measure attempts to correctly interpret how terms combine via their relationships in the ontological hierarchy. SSA uses these relationships to identify the most specific common ancestors between terms. We outline the set of cases in which terms can combine and associate partial order constraints with each case that order the specificity of terms. These cases form the basis for the SSA algorithm. The set of associated constraints also provide a set of principles that any improvement on our method should seek to satisfy.
Conclusion: We derive a measure of semantic similarity between annotations that exploits all available information without introducing assumptions about the nature of the ontology or data. We preserve the principles underlying instance based methods of semantic similarity of terms at the annotation level. As a result our measure better describes the information contained in annotations associated with gene products and as a result is better suited to characterizing and classifying gene products through their annotations.
Figures
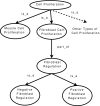
Figure 1
An Example of an Ontology of GO Terms. Nodes in the graph correspond to ontological terms. Edges correspond to relations between terms. Lower down terms in the diagram are descendants of terms higher up in the diagram if connected by an edge.
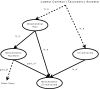
Figure 2
A Subset of GO Terms and Relations. An example of where the part_of relation plays an important role in interpreting annotations. If an annotation contains the term 'mitochondrial chromosome' then all other terms shown in the graph are redundant. The diagram also shows various cases that describe how terms relate to each other.
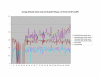
Figure 3
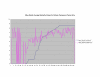
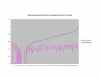
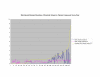
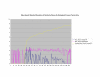
Normalized SSA Resnik vs Wang's Method vs Normalized Max Resnik. Values shown correspond to the average annotation similarity values between gene products with other gene products in the same pathway (taken from the SGD biochemical pathways database) and between gene products in a pathway with other gene products not found in the pathway.
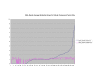
Figure 4
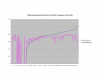
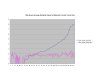
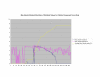
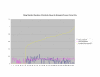
Average Pathway Similarity Values of Annotations Consisting only of Cellular Component Terms Using SSA Resnik. Average of SSA Resnik similarity values of gene products inside and outside a pathway.
Figure 5
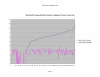
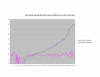
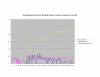
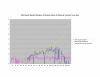
Average Pathway Similarity Values of Annotations Consisting only of Cellular Component Terms Using Max Resnik. Average of Max Resnik similarity values of gene products inside and outside a pathway.
Figure 6
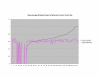
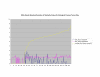
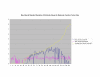
Average Pathway Similarity Values of Annotations Consisting only of Cellular Component Terms Using Wang's Method. Average of Wang's measure of similarity of gene products inside and outside a pathway.
Figure 7
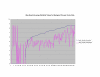
Average Pathway Similarity Values of Annotations Consisting only of Biological Process Terms Using SSA Resnik. Average of SSA Resnik similarity values of gene products inside and outside a pathway.
Figure 8
Average Pathway Similarity Values of Annotations Consisting only of Biological Process Terms Using Max Resnik. Average of Max Resnik similarity values of gene products inside and outside a pathway.
Figure 9
Average Pathway Similarity Values of Annotations Consisting only of Biological Process Terms Using Wang's Method. Average of Wang's measure of similarity of gene products inside and outside a pathway.
Figure 10
Average Pathway Similarity Values of Annotations Consisting only of Molecular Function Terms Using SSA Resnik. Average of SSA Resnik similarity values of gene products inside and outside a pathway.
Figure 11
Average Pathway Similarity Values of Annotations Consisting only of Molecular Function Terms Using Max Resnik. Average of Max Resnik similarity values of gene products inside and outside a pathway.
Figure 12
Average Pathway Similarity Values of Annotations Consisting only of Molecular Function Terms Using Wang's Method. Average of Wang's measure of similarity of gene products inside and outside a pathway.
Figure 13
Standard Deviation of Pathway Similarity Values of Annotations Consisting only of Cellular Component Terms Using SSA Resnik. Standard deviation of SSA Resnik similarity values of gene products inside and outside a pathway.
Figure 14
Standard Deviation of Pathway Similarity Values of Annotations Consisting only of Cellular Component Terms Using Max Resnik. Standard deviation of Max Resnik similarity values of gene products inside and outside a pathway.
Figure 15
Standard Deviation of Pathway Similarity Values of Annotations Consisting only of Cellular Component Terms Using Wang's Method. Standard deviation of values of Wang's measure of similarity of gene products inside and outside a pathway.
Figure 16
Standard Deviation of Pathway Similarity Values of Annotations Consisting only of Biological Process Terms Using SSA Resnik. Standard deviation of SSA Resnik similarity values of gene products inside and outside a pathway.
Figure 17
Standard Deviation of Pathway Similarity Values of Annotations Consisting only of Biological Process Terms Using Max Resnik. Standard deviation of Max Resnik similarity values of gene products inside and outside a pathway.
Figure 18
Standard Deviation of Pathway Similarity Values of Annotations Consisting only of Biological Process Terms Using Wang's Method. Standard deviation of values of Wang's measure of similarity of gene products inside and outside a pathway.
Figure 19
Standard Deviation of Pathway Similarity Values of Annotations Consisting only of Molecular Function Terms Using SSA Resnik. Standard deviation of SSA Resnik similarity values of gene products inside and outside a pathway.
Figure 20
Standard Deviation of Pathway Similarity Values of Annotations Consisting only of Molecular Function Terms Using Max Resnik. Standard deviation of Max Resnik similarity values of gene products inside and outside a pathway.
Figure 21
Standard Deviation of Pathway Similarity Values of Annotations Consisting only of Molecular Function Terms Using Wang's Method. Standard deviation of values of Wang's measure of similarity of gene products inside and outside a pathway.
Similar articles
- Semantic similarity in biomedical ontologies.
Pesquita C, Faria D, Falcão AO, Lord P, Couto FM. Pesquita C, et al. PLoS Comput Biol. 2009 Jul;5(7):e1000443. doi: 10.1371/journal.pcbi.1000443. Epub 2009 Jul 31. PLoS Comput Biol. 2009. PMID: 19649320 Free PMC article. Review. - GOSemSim: an R package for measuring semantic similarity among GO terms and gene products.
Yu G, Li F, Qin Y, Bo X, Wu Y, Wang S. Yu G, et al. Bioinformatics. 2010 Apr 1;26(7):976-8. doi: 10.1093/bioinformatics/btq064. Epub 2010 Feb 23. Bioinformatics. 2010. PMID: 20179076 - A new method to measure the semantic similarity of GO terms.
Wang JZ, Du Z, Payattakool R, Yu PS, Chen CF. Wang JZ, et al. Bioinformatics. 2007 May 15;23(10):1274-81. doi: 10.1093/bioinformatics/btm087. Epub 2007 Mar 7. Bioinformatics. 2007. PMID: 17344234 - Semantic similarity estimation in the biomedical domain: an ontology-based information-theoretic perspective.
Sánchez D, Batet M. Sánchez D, et al. J Biomed Inform. 2011 Oct;44(5):749-59. doi: 10.1016/j.jbi.2011.03.013. Epub 2011 Apr 2. J Biomed Inform. 2011. PMID: 21463704 - From ontology to semantic similarity: calculation of ontology-based semantic similarity.
Gan M, Dou X, Jiang R. Gan M, et al. ScientificWorldJournal. 2013;2013:793091. doi: 10.1155/2013/793091. Epub 2013 Feb 28. ScientificWorldJournal. 2013. PMID: 23533360 Free PMC article. Review.
Cited by
- Semantic similarity in biomedical ontologies.
Pesquita C, Faria D, Falcão AO, Lord P, Couto FM. Pesquita C, et al. PLoS Comput Biol. 2009 Jul;5(7):e1000443. doi: 10.1371/journal.pcbi.1000443. Epub 2009 Jul 31. PLoS Comput Biol. 2009. PMID: 19649320 Free PMC article. Review. - An improved approach to infer protein-protein interaction based on a hierarchical vector space model.
Zhang J, Jia K, Jia J, Qian Y. Zhang J, et al. BMC Bioinformatics. 2018 Apr 27;19(1):161. doi: 10.1186/s12859-018-2152-z. BMC Bioinformatics. 2018. PMID: 29699476 Free PMC article. - A shortest-path graph kernel for estimating gene product semantic similarity.
Alvarez MA, Qi X, Yan C. Alvarez MA, et al. J Biomed Semantics. 2011 Jul 29;2:3. doi: 10.1186/2041-1480-2-3. J Biomed Semantics. 2011. PMID: 21801410 Free PMC article. - Unveiling inter-embryo variability in spindle length over time: Towards quantitative phenotype analysis.
Le Cunff Y, Chesneau L, Pastezeur S, Pinson X, Soler N, Fairbrass D, Mercat B, Rodriguez-Garcia R, Alayan Z, Abdouni A, de Neidhardt G, Costes V, Anjubault M, Bouvrais H, Héligon C, Pécréaux J. Le Cunff Y, et al. PLoS Comput Biol. 2024 Sep 5;20(9):e1012330. doi: 10.1371/journal.pcbi.1012330. eCollection 2024 Sep. PLoS Comput Biol. 2024. PMID: 39236069 Free PMC article. - Identifying informative subsets of the Gene Ontology with information bottleneck methods.
Jin B, Lu X. Jin B, et al. Bioinformatics. 2010 Oct 1;26(19):2445-51. doi: 10.1093/bioinformatics/btq449. Epub 2010 Aug 11. Bioinformatics. 2010. PMID: 20702400 Free PMC article.
References
- Lord P, Stevens R, Brass A, Goble CA. Semantic Similarity Measures as Tools for Exploring the Gene Ontology. Pacific Symposium on Biocomputing. 2003;8:601–612. - PubMed
- Lord P, Stevens R, Brass A, Goble C. Investigating semantic similarity measures across the Gene Ontology: the relationship between sequence and annotation. Bioinformatics. 2003;19:1275–1283. - PubMed
- Sevilla J, Segura V, Podhorski A, Guruceaga JE Mato, Martinez-Cruz L, Corrales F, Rubio A. Correlation between gene expression and GO semantic similarity. IEEE/ACM Transactions on Computational Biology and Bioinformatics. 2005;2:330–338. - PubMed
- Couto FM, Silva MJ, Coutinho PM. Measuring semantic similarity between Gene Ontology terms, Data and Knowledge Engineering. Business Process Management – Where business processes and web services meet. 2007;61:137–152.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials