Proteomic changes associated with diabetes in the BB-DP rat - PubMed (original) (raw)
Proteomic changes associated with diabetes in the BB-DP rat
D Thor Johnson et al. Am J Physiol Endocrinol Metab. 2009 Mar.
Erratum in
- Am J Physiol Endocrinol Metab. 2010 Apr;298(4):E895
Abstract
These studies were structured with the aim of utilizing emerging technologies in two-dimensional (2D) gel electrophoresis and mass spectrometry to evaluate protein expression changes associated with type 1 diabetes. We reasoned that a broad examination of diabetic tissues at the protein level might open up novel avenues of investigation of the metabolic and signaling pathways that are adversely affected in type 1 diabetes. This study compared the protein expression of the liver, heart, and skeletal muscle of diabetes-prone rats and matched control rats by semiquantitative liquid chromatography-mass spectrometry and differential in-gel 2D gel electrophoresis. Differential expression of 341 proteins in liver, 43 in heart, and 9 (2D gel only) in skeletal muscle was detected. These data were assembled into the relevant metabolic pathways affected primarily in liver. Multiple covalent modifications were also apparent in 2D gel analysis. Several new hypotheses were generated by these data, including mechanisms of net cytosolic protein oxidation, formaldehyde generation by the methionine cycle, and inhibition of carbon substrate oxidation via reduction in citrate synthase and short-chain acyl-CoA dehydrogenase.
Figures
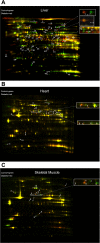
Fig. 1.
Two-dimensional differential gel electrophoresis of diabetic tissues. Overlay of diabetic vs. control tissue. A: liver. B: heart. C: skeletal muscle. Protein identifications follow. Liver: 1) aldehyde dehydrogenase 1; 2) sarcosine dehydrogenase; 3) dimethylglycine dehydrogenase; 4) serotransferrin precursor; 5) mitochondrial aconitase; 6) long-chain-fatty acid-CoA ligase; 7) urocanase; 8) glucokinase regulatory protein; 9) phospho_enol_pyruvate caboxykinase; 10) succinate dehydrogenase flavoprotein; 11); stress 70 protein; 12) protein disulfide-isomerase; 13) 78-kDa glucose-regulated protein; 14) protein disulfide isomerase A3; 15) pyruvate kinase isozymes R/L; 16) phosphoglucomutase 1; 17) carbamoyl-phosphate synthase II; 18) liver carboxylesterase 10; 19) serine protease inhibitor A3k; 20) glycerol kinase; 21) serine protease inhibitor A3L; 22) _S_-adenosylmethionine synthetase; 23) argininosuccinate lyase; 24) β-ureidopropionase; 25) cysteine sulfinic acid decarboxylase; 26) hydroxymethylglutaryl-CoA synthase; 27) fumarate hydratase; 28) argininosuccinate synthase; 29) glutamate oxaloacetate transaminase 1; 30) apolipoprotein A-IV; 31) regucalcin; 32) 3-oxo-5-β-steroid 4-dehydrogenase; 33) arginase I; 34) _N_-hydroxyarylamine sulfotransferase; 35) cytosolicmalate dehydrogenase; 36) estrogen sulfotransferase isoform 2; 37) glutamate-cysteine ligase modifier subunit; 38) prohibitin; 39) proteasome activator complex subunit 2; 40) phenazine biosynthesis-like domain-containing protein; 41) Δ3;5-Δ2;4-dienoyl-CoA isomerase; 42) homogentisate oxygenase; 43) glycoprotein gC1qBP; 44) catechol _O_-methyltransferase, membrane-bound form; 45) major urinary protein; 46) pterin-4-α-carbinolamine dehydratase; 47) D-dopachrome tautomerase; 48) fatty acid-binding protein. Skeletal muscle: 1) glycogen phosphorylase, muscle form; 2) serotransferrin, splice isoform 2; 3) serum albumin; 4) tripartite motif-containing protein 72; 5) enolase 1α; 6) fumerase; 8) apolipoprotein A-IV; 9) apolipoprotein A-I; 10) atrial natriuretic factor; 11) ATP synthase delta chain. Heart: 1) LIM domain-binding protein 3; 2) creatine kinase; 3) ATP synthesis subunit β; 4) tripartite motif-containing protein 72; 5) α-enolase; 6) succinyl-CoAligase (ADP-forming)-β; 7) isocitrate dehydrogenase [NAD]-α; 8) isocitrate dehydrogenase [NAD]-α; 9) ATP synthase delta chain; 10) serum albumin.
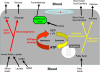
Fig. 2.
Citric acid cycle. Proteins of the citric acid cycle detected in this screen. In all figures, proteins upregulated by ≥20% are denoted by a thick line, proteins downregulated by ≥20% by a dotted line, proteins within 20% in expression by a thin solid line. Gray line indicates that the protein was not detected. Symbol and protein identification: citrate synthase (CS); aconitase (AC); NAD+ isocitrate dehydrogenase (ICDH); 2-oxoglutarate dehydrogenase (2-ODH); succinyl-CoA ligase (SCoAL); succinate dehydrogenase (SDH); fumerate hydratase (FH); malate dehydrogenase (MDH); hydroxyacid-oxoacid transhydrogenase (HOT; known in the literature before 2006 as alcohol dehydrogenase) iron containing 1; succinate semialdehyde dehydrogenase (SSDH); aminobutyrate transaminase (ABT); glutamate oxaloacetate transaminase 1 (GOT). Electron transport chain. Comparison of protein expression of diabetic vs. control in components of the electron transport chain. Several proteins were grouped into complexes as outlined below. Complex 1: NADH dehydrogenase (ubiquinone) 1α subcomplex 9. Electron transfer flavoprotein: electron transfer flavoprotein-α; electron-transferring flavoprotein dehydrogenase. Complex 2: succinate dehydrogenase complex subunit B. Complex 3: ubiquinol-cytochrome c reductase core protein 1; ubiquinol-cytochrome c reductase complex core protein 2. Complex 4: cytochrome c oxidase polypeptide Va; cytochrome c oxidase subunit IV isoform 1; cytochrome c oxidase polypeptide Via. F1F0ATPase: ATP synthase β-chain; H+-ATP synthase-α; ATP synthase coupling factor 6; ATP synthase H+ transporting mitochondrial F1 complex-γ; ATP synthase B; ATP synthase subunit d; ATP synthase oligomycin sensitivity conferral protein. Others: ADP, ATP carrier protein (ANT); phosphate transport protein (PT). Fatty acid oxidaion. Symbol and protein identification: carnitine palmitoyltransferase I (CPT1); carnitine palmitoyltransferase II (CPT2); acyl-CoA dehydrogenase long-chain specific (LCAD); acyl-CoA dehydrogenase medium-chain specific (MCAD); acyl-CoA dehydrogenase short-chain specific (SCAD); acyl-CoA dehydrogenase very long-chain specific (VLCAD); hydroxymethylglutaryl-CoA synthase (HMGCoAS); hydroxymethylglutaryl-CoA lyase (HMGCoAL); 3-ketoacyl-CoA thiolase (thiolase); 2,4-dienoyl-CoA reductase (DCoAR); Δ3,5-Δ2,4-dienoyl-CoA isomerase (DCoAI); 2-_trans_-enoyl-CoA isomerase (ECo-AI); trifuctional enzyme (TFE); β-hydroxybutyrate dehydrogenase (BHBH).
Fig. 3.
Gluconeogenesis/glycolysis. Protein identifications as follows: serine hydratase (SH); glucose-6-phosphate isomerase (PGI); triosephosphate isomerase 1 (TPI); phosphoglycerate mutase 1 (PM); phosphoglycerate kinase 1 (PGK); enolase 1α (EN); phospho_enol_pyruvate carboxykinase (PEPCK); pyruvate carboxylase (PC); fructose-1,6-biphosphatase 1 (F-1,6-B); malate dehydrogenase mitochondrial (mMD); malate dehydrogenasecytosolic (cMD); aspartate transaminase cytosolic (cAT); aspartate transaminase mitochondrial (mAT); glucose phophatase (GP); phosphoglucomutase 1(PGM); glucose-6-phosphate isomerase (PGI); phosphoglycerate mutase 1 (PGM); pyruvate kinase M2 (PK); phosphoglycerate kinase 1 (PGK); glyceraldehyde-3-phosphate dehydrogenase (G-3PD); aldolase B (FBPA); hexokinase (HK); phosphofructokinase 1 (PFK1); alanine aminotransferase (AAT).
Fig. 4.
Urea cycle. Elements of the urea cycle isolated from diabetic and control liver. Symbol and protein identification: glutamate dehydrogenase I (GDH1); carbamoyl phosphate synthase (CPS); arginase I (Arg 1); ornithine aminotransferase (OAT); ornithine carbamoyltransferase (OCT); arginosuccinate synthase (ASS); arginosuccinate lyase (ASL). ROS metabolism. Elements of ROS metabolism. Symbol identification: glutathione peroxidase (GPX-1); Mn superoxide dismutase (Mn-SOD) Cu superoxide dismutase (Cu-SOD); cytosolic peroxiredoxins 6,1,4 (Prx-6,1,4); mitochondrial peroxiredoxin 5 (Prx-5), catalase. Methionine cycle. Elements of the methionine cycle. Symbol indentification: methionine adenosyltransferase (MAT); betaine-homocysteine _S_-methyltransferase (BHMT); glycine methyltransferase (GNMT); catechol _O_-methyltransferase (COMT); guanidinoacetate methyltransferase (GAMT); _S_-adenosylhomocysteine hydrolase (SAHH); choline oxidoreductase (CO); betaine aldehyde dehydrogenase (BAD); electron transfer flavoprotein (ETF); sarcosine dehydrogenase (SaDH); dimethylglycine dehydrogenase (DMDH); serine hydroxymethyl transferase (SHMT); tetrahydrofolate (THF).
Fig. 5.
Overall metabolic summary of focus of the liver in the diabetic state. Yellow areas concern the conversion of energy into ATP; red arrows represent upregulated ATP hydrolysis reactions generating work.
Similar articles
- Different islet protein expression profiles during spontaneous diabetes development vs. allograft rejection in BB-DP rats.
Christensen UB, Larsen PM, Fey S, Karlsen AE, Pociot F, Nerup J, Sparre T. Christensen UB, et al. Autoimmunity. 2006 Jun;39(4):315-21. doi: 10.1080/08916930600648269. Autoimmunity. 2006. PMID: 16891220 - A combined 1H-NMR spectroscopy- and mass spectrometry-based metabolomic study of the PPAR-alpha null mutant mouse defines profound systemic changes in metabolism linked to the metabolic syndrome.
Atherton HJ, Bailey NJ, Zhang W, Taylor J, Major H, Shockcor J, Clarke K, Griffin JL. Atherton HJ, et al. Physiol Genomics. 2006 Oct 11;27(2):178-86. doi: 10.1152/physiolgenomics.00060.2006. Epub 2006 Jul 25. Physiol Genomics. 2006. PMID: 16868074 - Differential expression and glycative damage affect specific mitochondrial proteins with aging in rat liver.
Bakala H, Ladouce R, Baraibar MA, Friguet B. Bakala H, et al. Biochim Biophys Acta. 2013 Dec;1832(12):2057-67. doi: 10.1016/j.bbadis.2013.07.015. Epub 2013 Jul 30. Biochim Biophys Acta. 2013. PMID: 23906978 - Regulatory role for the arginine-nitric oxide pathway in metabolism of energy substrates.
Jobgen WS, Fried SK, Fu WJ, Meininger CJ, Wu G. Jobgen WS, et al. J Nutr Biochem. 2006 Sep;17(9):571-88. doi: 10.1016/j.jnutbio.2005.12.001. Epub 2006 Jan 9. J Nutr Biochem. 2006. PMID: 16524713 Review. - [Regulation of biosynthesis and oxidation of fatty acids and glycerides at the cellular level in mammalian tissues].
Alimova EK, Astvatsatur'ian AT, Zharov LV. Alimova EK, et al. Vopr Med Khim. 1974 Sep-Oct;20(5):451-62. Vopr Med Khim. 1974. PMID: 4156442 Review. Russian. No abstract available.
Cited by
- What can we learn about cardioprotection from the cardiac mitochondrial proteome?
Gucek M, Murphy E. Gucek M, et al. Cardiovasc Res. 2010 Nov 1;88(2):211-8. doi: 10.1093/cvr/cvq277. Epub 2010 Aug 30. Cardiovasc Res. 2010. PMID: 20805096 Free PMC article. Review. - Hepatic proteomic analysis revealed altered metabolic pathways in insulin resistant Akt1(+/-)/Akt2(-/-) mice.
Pedersen BA, Wang W, Taylor JF, Khattab OS, Chen YH, Edwards RA, Yazdi PG, Wang PH. Pedersen BA, et al. Metabolism. 2015 Dec;64(12):1694-703. doi: 10.1016/j.metabol.2015.09.008. Epub 2015 Sep 12. Metabolism. 2015. PMID: 26455965 Free PMC article. - Proteomic analysis of glycated proteins from streptozotocin-induced diabetic rat kidney.
Chougale AD, Bhat SP, Bhujbal SV, Zambare MR, Puntambekar S, Somani RS, Boppana R, Giri AP, Kulkarni MJ. Chougale AD, et al. Mol Biotechnol. 2012 Jan;50(1):28-38. doi: 10.1007/s12033-011-9409-3. Mol Biotechnol. 2012. PMID: 21516357 - The mitochondrial proteome: a dynamic functional program in tissues and disease states.
Balaban RS. Balaban RS. Environ Mol Mutagen. 2010 Jun;51(5):352-9. doi: 10.1002/em.20574. Environ Mol Mutagen. 2010. PMID: 20544878 Free PMC article. Review. - Living without creatine: unchanged exercise capacity and response to chronic myocardial infarction in creatine-deficient mice.
Lygate CA, Aksentijevic D, Dawson D, ten Hove M, Phillips D, de Bono JP, Medway DJ, Sebag-Montefiore L, Hunyor I, Channon KM, Clarke K, Zervou S, Watkins H, Balaban RS, Neubauer S. Lygate CA, et al. Circ Res. 2013 Mar 15;112(6):945-55. doi: 10.1161/CIRCRESAHA.112.300725. Epub 2013 Jan 16. Circ Res. 2013. PMID: 23325497 Free PMC article.
References
- Aerts L, Van Assche FA. Low taurine, gamma-aminobutyric acid and carnosine levels in plasma of diabetic pregnant rats: consequences for the offspring. J Perinat Med 29: 81–84, 2001. - PubMed
- Almdal TP, Jensen T, Vilstrup H. Increased hepatic efficacy of urea synthesis from alanine in insulin-dependent diabetes mellitus. Eur J Clin Invest 20: 29–34, 1990. - PubMed
- Almdal TP, Petersen KF, Hansen BA, Vilstrup H. Increased capacity of urea synthesis in streptozotocin diabetes in rats. Diabetologia 29: 812–816, 1986. - PubMed
- Appel MC, Like AA, Rossini AA, Carp DB, Miller TB Jr. Hepatic carbohydrate metabolism in the spontaneously diabetic Bio-Breeding Worcester rat. Am J Physiol Endocrinol Metab 240: E83–E87, 1981. - PubMed
- Barreto EO, Riederer I, Arantes AC, Carvalho VF, Farias-Filho FA, Cordeiro RS, Martins MA, Savino W, Silva PM. Thymus involution in alloxan diabetes: analysis of mast cells. Mem Inst Oswaldo Cruz 100, Suppl 1: 127–130, 2005. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical