Detection of genetic association and a functional polymorphism of dynamin 1 gene with nicotine dependence in European and African Americans - PubMed (original) (raw)
Detection of genetic association and a functional polymorphism of dynamin 1 gene with nicotine dependence in European and African Americans
Qing Xu et al. Neuropsychopharmacology. 2009 Apr.
Abstract
Although it has been documented that dynamin 1 gene (DNM1) is significantly modulated by nicotine in animal models, its association with nicotine dependence (ND) in human population remained to be unexplored. To determine whether DNM1 is associated with ND, in this study, we genotyped seven single-nucleotide polymorphisms (SNPs) within this gene in 602 nuclear families of either African-American (AA) or European-American (EA) origin. Individual SNP-based association analysis revealed a significant association of SNP rs3003609 with smoking quantity (SQ; P=0.0031) and Heaviness of Smoking Index (HSI; P=0.0042) in the EA sample. Furthermore, our haplotype-based association analyses indicated that haplotypes T-G-T, formed by rs2502731-rs2229917-rs3003609 (at a frequency of 54%), G-T-A, formed by rs2229917-rs3003609-rs16930313 (at a frequency of 52%), and T-A-G, formed by rs3003609-rs16930313-rs7022174 (at a frequency of 52%) are significantly associated with SQ (Z=-2.44 to -2.92; P=0.015-0.0055) and HSI (Z=-2.52 to -2.67; P=0.012-0.0076) in the EA sample. In the AA sample, another haplotype, G-T-A, formed by rs7875406-rs2502731-rs2229917, at a frequency of 12% was significantly associated with SQ (Z=-2.58; P=0.0098). Finally, by using in vitro gene expression assays, we demonstrated that the T allele of rs3003609 in the exon 9 of DNM1 significantly decreases the expression of DNM1, by 27.1% at the mRNA and 22.0% at the protein level, suggesting that rs3003609 represents a functional polymorphism affecting DNM1 expression and may partly contributed to the observed association of the gene with ND in our samples. Taken together, our findings indicate that DNM1 is likely involved in the etiology of ND and represents a plausible candidate for further investigation in independent samples.
Conflict of interest statement
Disclosure/Conflict of Interest
QX, WH, TJP, and JZM have no conflicts of interest to declare. MDL disclosed consulting fees on a NIDA-sponsored research project from Information Management Consultants, Inc.
Figures
Figure 1
Haploview-generated LD maps of the seven SNPs within DNM1 in the EA and AA samples. Regions of high LD (D′ = 1 and LD >2) are shown in dark gray. Markers with LD (0.21 < D′ < 1 and LOD >2) are shown in dark through light gray, with the color intensity decreasing with decreasing D′ values. Regions of low LD and low LOD scores (LOD <2) are shown in white. The number within each box indicates the D′ statistic between the two SNPs. Haplotype blocks in the two samples were produced by the HaploView program using the block definitions proposed by Grabiel et al.
Figure 2
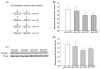
Expression analyses of allele combinations of SNPs rs2229917 at exon 4 and rs3003609 at exon 9. (A) Illustration of four plasmids constructed for dynamin 1 expression analysis. (B) Statistical analysis of real-time RT-PCR data for the four plasmids. 18S RNA was used to normalize expression of each DNM1 construct. Compared with pE4G-E9C, pE4G-E9T and pE4A-E9T showed lower expression of dynamin 1, by 27.1% and 26.8%, respectively. No significant difference was detected between constructs pE4G-E9C and pE4A-E9C. (C) Representative Western blotting images for dynamin 1 and tubulin. Tubulin was used to normalize expression of each DNM1 construct. (D) Statistical analysis of the protein expression levels of four DNM1 constructs. Similar to the mRNA expression data in (C), pE4G-E9T and pE4A-E9T showed decreased dynamin 1 protein expression, by 29.2% and 22.2%, respectively, compared with pE4G-E9C. Data in Figures B and D are given as means ± S.E.M. (* P <0.05; n = 3/group).
Figure 3
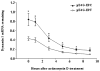
Comparison of mRNA stability of DNM1 from pE4G-E9C and pE4G-E9T constructs. Measured half-life of mRNA was about 3.2 hours for the former and 3.6 hours for the latter. No significant difference in the half-life of DNM1 mRNA was found in the two constructs. Data are given as means ± S.E.M. (* P <0.05; n = 3/group).
Similar articles
- Fine mapping of a linkage region on chromosome 17p13 reveals that GABARAP and DLG4 are associated with vulnerability to nicotine dependence in European-Americans.
Lou XY, Ma JZ, Sun D, Payne TJ, Li MD. Lou XY, et al. Hum Mol Genet. 2007 Jan 15;16(2):142-53. doi: 10.1093/hmg/ddl450. Epub 2006 Dec 12. Hum Mol Genet. 2007. PMID: 17164261 - Association of amyloid precursor protein-binding protein, family B, member 1 with nicotine dependence in African and European American smokers.
Chen GB, Payne TJ, Lou XY, Ma JZ, Zhu J, Li MD. Chen GB, et al. Hum Genet. 2008 Nov;124(4):393-8. doi: 10.1007/s00439-008-0558-9. Epub 2008 Sep 7. Hum Genet. 2008. PMID: 18777128 Free PMC article. - Linkage and association studies in African- and Caucasian-American populations demonstrate that SHC3 is a novel susceptibility locus for nicotine dependence.
Li MD, Sun D, Lou XY, Beuten J, Payne TJ, Ma JZ. Li MD, et al. Mol Psychiatry. 2007 May;12(5):462-73. doi: 10.1038/sj.mp.4001933. Epub 2006 Dec 19. Mol Psychiatry. 2007. PMID: 17179996 - Significant association of the neurexin-1 gene (NRXN1) with nicotine dependence in European- and African-American smokers.
Nussbaum J, Xu Q, Payne TJ, Ma JZ, Huang W, Gelernter J, Li MD. Nussbaum J, et al. Hum Mol Genet. 2008 Jun 1;17(11):1569-77. doi: 10.1093/hmg/ddn044. Epub 2008 Feb 11. Hum Mol Genet. 2008. PMID: 18270208 Free PMC article.
Cited by
- Non-coding RNAs--novel targets in neurotoxicity.
Tal TL, Tanguay RL. Tal TL, et al. Neurotoxicology. 2012 Jun;33(3):530-44. doi: 10.1016/j.neuro.2012.02.013. Epub 2012 Feb 27. Neurotoxicology. 2012. PMID: 22394481 Free PMC article. Review. - Nicotine modulates expression of dynamin 1 in rat brain and SH-SY5Y cells.
Xu Q, Li MD. Xu Q, et al. Neurosci Lett. 2011 Feb 11;489(3):168-71. doi: 10.1016/j.neulet.2010.12.009. Epub 2010 Dec 13. Neurosci Lett. 2011. PMID: 21159320 Free PMC article. - MicroRNAs in addiction: adaptation's middlemen?
Li MD, van der Vaart AD. Li MD, et al. Mol Psychiatry. 2011 Dec;16(12):1159-68. doi: 10.1038/mp.2011.58. Epub 2011 May 24. Mol Psychiatry. 2011. PMID: 21606928 Free PMC article. Review. - Common and unique biological pathways associated with smoking initiation/progression, nicotine dependence, and smoking cessation.
Wang J, Li MD. Wang J, et al. Neuropsychopharmacology. 2010 Feb;35(3):702-19. doi: 10.1038/npp.2009.178. Epub 2009 Nov 4. Neuropsychopharmacology. 2010. PMID: 19890259 Free PMC article. - The contribution of rare and common variants in 30 genes to risk nicotine dependence.
Yang J, Wang S, Yang Z, Hodgkinson CA, Iarikova P, Ma JZ, Payne TJ, Goldman D, Li MD. Yang J, et al. Mol Psychiatry. 2015 Nov;20(11):1467-78. doi: 10.1038/mp.2014.156. Epub 2014 Dec 2. Mol Psychiatry. 2015. PMID: 25450229 Free PMC article.
References
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21(2):263–265. - PubMed
- Benowitz NL, Perez-Stable EJ, Fong I, Modin G, Herrera B, Jacob P., 3rd Ethnic differences in N-glucuronidation of nicotine and cotinine. J Pharmacol Exp Ther. 1999;291(3):1196–1203. - PubMed
- Carmelli D, Swan GE, Robinette D, Fabsitz R. Genetic influence on smoking--a study of male twins. N Engl J Med. 1992;327(12):829–833. - PubMed
- Ferguson SM, Brasnjo G, Hayashi M, Wolfel M, Collesi C, Giovedi S, et al. A selective activity-dependent requirement for dynamin 1 in synaptic vesicle endocytosis. Science. 2007;316(5824):570–574. - PubMed
- Frank S, Gaume B, Bergmann-Leitner ES, Leitner WW, Robert EG, Catez F, et al. The role of dynamin-related protein 1, a mediator of mitochondrial fission, in apoptosis. Dev Cell. 2001;1(4):515–525. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources