The diploid genome sequence of an Asian individual - PubMed (original) (raw)
Comparative Study
. 2008 Nov 6;456(7218):60-5.
doi: 10.1038/nature07484.
Wei Wang, Ruiqiang Li, Yingrui Li, Geng Tian, Laurie Goodman, Wei Fan, Junqing Zhang, Jun Li, Juanbin Zhang, Yiran Guo, Binxiao Feng, Heng Li, Yao Lu, Xiaodong Fang, Huiqing Liang, Zhenglin Du, Dong Li, Yiqing Zhao, Yujie Hu, Zhenzhen Yang, Hancheng Zheng, Ines Hellmann, Michael Inouye, John Pool, Xin Yi, Jing Zhao, Jinjie Duan, Yan Zhou, Junjie Qin, Lijia Ma, Guoqing Li, Zhentao Yang, Guojie Zhang, Bin Yang, Chang Yu, Fang Liang, Wenjie Li, Shaochuan Li, Dawei Li, Peixiang Ni, Jue Ruan, Qibin Li, Hongmei Zhu, Dongyuan Liu, Zhike Lu, Ning Li, Guangwu Guo, Jianguo Zhang, Jia Ye, Lin Fang, Qin Hao, Quan Chen, Yu Liang, Yeyang Su, A San, Cuo Ping, Shuang Yang, Fang Chen, Li Li, Ke Zhou, Hongkun Zheng, Yuanyuan Ren, Ling Yang, Yang Gao, Guohua Yang, Zhuo Li, Xiaoli Feng, Karsten Kristiansen, Gane Ka-Shu Wong, Rasmus Nielsen, Richard Durbin, Lars Bolund, Xiuqing Zhang, Songgang Li, Huanming Yang, Jian Wang
Affiliations
- PMID: 18987735
- PMCID: PMC2716080
- DOI: 10.1038/nature07484
Comparative Study
The diploid genome sequence of an Asian individual
Jun Wang et al. Nature. 2008.
Abstract
Here we present the first diploid genome sequence of an Asian individual. The genome was sequenced to 36-fold average coverage using massively parallel sequencing technology. We aligned the short reads onto the NCBI human reference genome to 99.97% coverage, and guided by the reference genome, we used uniquely mapped reads to assemble a high-quality consensus sequence for 92% of the Asian individual's genome. We identified approximately 3 million single-nucleotide polymorphisms (SNPs) inside this region, of which 13.6% were not in the dbSNP database. Genotyping analysis showed that SNP identification had high accuracy and consistency, indicating the high sequence quality of this assembly. We also carried out heterozygote phasing and haplotype prediction against HapMap CHB and JPT haplotypes (Chinese and Japanese, respectively), sequence comparison with the two available individual genomes (J. D. Watson and J. C. Venter), and structural variation identification. These variations were considered for their potential biological impact. Our sequence data and analyses demonstrate the potential usefulness of next-generation sequencing technologies for personal genomics.
Figures
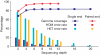
Figure 1. The percentage of detected SNPs (a) and small indels (b) that overlap with SNPs and small indels in the dbSNP database (http://www.ncbi.nlm.nih.gov/SNP/, build 128)
The dbSNP alleles were separated into validated and non-validated SNPs, and the detected SNPs that were not present in dbSNP were classified as novel.
Figure 2. Genome coverage of the assembled consensus sequence and the accuracy of SNP detection as a function of sequencing depth
Analyses were carried out on human chromosome 12, and subsets of reads from all mapped 22.5× single-end and 13.5× paired-end reads were randomly extracted from areas of different average depth. The same method and filtering threshold (Q20) was used for SNP detection over different sequencing depths. The error rate for SNP calling—the sum of ‘over call’, ‘under call’ and ‘misses’ rate (see Supplementary Information)—was separated into heterozygotes (HET) and homozygotes (HOM), and was validated against the Illumina 1M genotyping alleles.
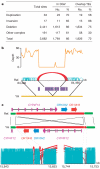
Figure 3. Summary of structural variations
a, Abundance of each class of structural variation. The overlap with known structural variations in the DGV (
http://projects.tcag.ca/variation/
) and with transposons (transposable elements, TEs) was calculated. About 34% of our identified structural variations are novel (having less than 10% of a portion of the YH structural variations overlapping with structural variations in the DGV). Transposable elements are a major component of the identified deletions, with Alus and LINEs involved in 49% and 34% of the deletions, respectively. b, An example of a deletion of a transposon complex on YH chromosome 1. The sequencing depth by both single-end and paired-end reads are shown. Normally aligned paired-end reads are shown in green, whereas abnormally aligned paired-end reads, which have unexpected long insert sizes or an incorrect orientation relationship, are shown in red. c, An example of an inversion on YH chromosome 19. Local assembly showed that a 102,405-bp fragment was inverted and reinserted in the genome. There are three genes in this sequence fragment, and the last exon of gene CYP4F12 was destroyed by this inversion event.
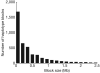
Figure 4. Size distribution of predicted haplotype blocks of autosomes
Haplotypes were constructed using PHASE software with the 700,300 autosomal heterozygous SNPs that overlapped with the CHB/JPT genotypes from the HapMap phase II data.
Comment in
- Human genetics: Individual genomes diversify.
Levy S, Strausberg RL. Levy S, et al. Nature. 2008 Nov 6;456(7218):49-51. doi: 10.1038/456049a. Nature. 2008. PMID: 18987731 No abstract available.
Similar articles
- De novo assembly and phasing of a Korean human genome.
Seo JS, Rhie A, Kim J, Lee S, Sohn MH, Kim CU, Hastie A, Cao H, Yun JY, Kim J, Kuk J, Park GH, Kim J, Ryu H, Kim J, Roh M, Baek J, Hunkapiller MW, Korlach J, Shin JY, Kim C. Seo JS, et al. Nature. 2016 Oct 13;538(7624):243-247. doi: 10.1038/nature20098. Epub 2016 Oct 5. Nature. 2016. PMID: 27706134 - The complete genome of an individual by massively parallel DNA sequencing.
Wheeler DA, Srinivasan M, Egholm M, Shen Y, Chen L, McGuire A, He W, Chen YJ, Makhijani V, Roth GT, Gomes X, Tartaro K, Niazi F, Turcotte CL, Irzyk GP, Lupski JR, Chinault C, Song XZ, Liu Y, Yuan Y, Nazareth L, Qin X, Muzny DM, Margulies M, Weinstock GM, Gibbs RA, Rothberg JM. Wheeler DA, et al. Nature. 2008 Apr 17;452(7189):872-6. doi: 10.1038/nature06884. Nature. 2008. PMID: 18421352 - Haplotype-resolved genome sequencing of a Gujarati Indian individual.
Kitzman JO, Mackenzie AP, Adey A, Hiatt JB, Patwardhan RP, Sudmant PH, Ng SB, Alkan C, Qiu R, Eichler EE, Shendure J. Kitzman JO, et al. Nat Biotechnol. 2011 Jan;29(1):59-63. doi: 10.1038/nbt.1740. Epub 2010 Dec 19. Nat Biotechnol. 2011. PMID: 21170042 Free PMC article. - Haplotyping-Assisted Diploid Assembly and Variant Detection with Linked Reads.
Hu Y, Yang C, Zhang L, Zhou X. Hu Y, et al. Methods Mol Biol. 2023;2590:161-182. doi: 10.1007/978-1-0716-2819-5_11. Methods Mol Biol. 2023. PMID: 36335499 Review. - A case study of the utility of the HapMap database for pharmacogenomic haplotype analysis in the Taiwanese population.
Lin E, Hwang Y, Tzeng CM. Lin E, et al. Mol Diagn Ther. 2006;10(6):367-70. doi: 10.1007/BF03256213. Mol Diagn Ther. 2006. PMID: 17154653 Review.
Cited by
- Deep whole-genome sequencing of 100 southeast Asian Malays.
Wong LP, Ong RT, Poh WT, Liu X, Chen P, Li R, Lam KK, Pillai NE, Sim KS, Xu H, Sim NL, Teo SM, Foo JN, Tan LW, Lim Y, Koo SH, Gan LS, Cheng CY, Wee S, Yap EP, Ng PC, Lim WY, Soong R, Wenk MR, Aung T, Wong TY, Khor CC, Little P, Chia KS, Teo YY. Wong LP, et al. Am J Hum Genet. 2013 Jan 10;92(1):52-66. doi: 10.1016/j.ajhg.2012.12.005. Epub 2013 Jan 3. Am J Hum Genet. 2013. PMID: 23290073 Free PMC article. - Development of InDel markers for Brassica rapa based on whole-genome re-sequencing.
Liu B, Wang Y, Zhai W, Deng J, Wang H, Cui Y, Cheng F, Wang X, Wu J. Liu B, et al. Theor Appl Genet. 2013 Jan;126(1):231-9. doi: 10.1007/s00122-012-1976-6. Epub 2012 Sep 13. Theor Appl Genet. 2013. PMID: 22972202 - Personal genomes, participatory genomics and the anonymity-privacy conundrum.
Scaria V. Scaria V. J Genet. 2014 Dec;93(3):917-20. doi: 10.1007/s12041-014-0451-3. J Genet. 2014. PMID: 25572254 No abstract available. - Whole genome sequences of a male and female supercentenarian, ages greater than 114 years.
Sebastiani P, Riva A, Montano M, Pham P, Torkamani A, Scherba E, Benson G, Milton JN, Baldwin CT, Andersen S, Schork NJ, Steinberg MH, Perls TT. Sebastiani P, et al. Front Genet. 2012 Jan 3;2:90. doi: 10.3389/fgene.2011.00090. eCollection 2011. Front Genet. 2012. PMID: 22303384 Free PMC article. - Rainbow: a tool for large-scale whole-genome sequencing data analysis using cloud computing.
Zhao S, Prenger K, Smith L, Messina T, Fan H, Jaeger E, Stephens S. Zhao S, et al. BMC Genomics. 2013 Jun 27;14:425. doi: 10.1186/1471-2164-14-425. BMC Genomics. 2013. PMID: 23802613 Free PMC article.
References
- International Human Genome Sequencing Consortium Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. - PubMed
- Venter JC, et al. The sequence of the human genome. Science. 2001;291:1304–1351. - PubMed
- Wheeler DA, et al. The complete genome of an individual by massively parallel DNA sequencing. Nature. 2008;452:872–876. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources