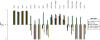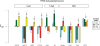High-throughput chromatin information enables accurate tissue-specific prediction of transcription factor binding sites - PubMed (original) (raw)
High-throughput chromatin information enables accurate tissue-specific prediction of transcription factor binding sites
Tom Whitington et al. Nucleic Acids Res. 2009 Jan.
Abstract
In silico prediction of transcription factor binding sites (TFBSs) is central to the task of gene regulatory network elucidation. Genomic DNA sequence information provides a basis for these predictions, due to the sequence specificity of TF-binding events. However, DNA sequence alone is an impoverished source of information for the task of TFBS prediction in eukaryotes, as additional factors, such as chromatin structure regulate binding events. We show that incorporating high-throughput chromatin modification estimates can greatly improve the accuracy of in silico prediction of in vivo binding for a wide range of TFs in human and mouse. This improvement is superior to the improvement gained by equivalent use of either transcription start site proximity or phylogenetic conservation information. Importantly, predictions made with the use of chromatin structure information are tissue specific. This result supports the biological hypothesis that chromatin modulates TF binding to produce tissue-specific binding profiles in higher eukaryotes, and suggests that the use of chromatin modification information can lead to accurate tissue-specific transcriptional regulatory network elucidation.
Figures
Figure 1.
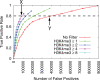
Improvement in E2F1 TFBS prediction by H3K4me3 signal filtering. ROC-like plot shows the TP rate versus the actual number of FPs. Error bars indicate standard error. The TF gold-standard and H3K4me3 data are each derived from mouse ES cells. This figure also serves to illustrate calculation of the ‘best relative FP improvement statistic’, (Is), defined in the Methods section.
Figure 2.
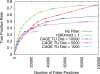
Comparison of H3K4me3 and TSS proximity filter performance for Klf4 TFBS prediction. ROC-like plot shows the TP rate versus the actual number of FPs. Error bars indicate standard error. The TF gold-standard and H3K4me3 data are each derived from mouse ES cells. A subset of all CAGE thresholds are presented for clarity.
Figure 3.
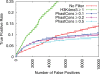
Comparison of H3K4me3 and phastCons filter performance for nMyc TFBS prediction. ROC-like plot shows the TP rate versus the actual number of FPs. Error bars indicate standard error. The TF gold-standard and H3K4me3 datasets are each derived from mouse ES cells. PhastCons filter performance for the other mouse TFs considered is similar to performance shown here for nMyc, as the optimal phastCons filter never outperforms the optimal H3K4me3 filter, for any TF or sensitivity level.
Figure 4.
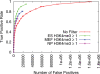
Tissue specificity of cMyc TFBS predictions made with H3K4me3 filter. ROC-like plot shows the TP rate versus the actual number of FPs. Error bars indicate standard error. The TF gold-standard data are each derived from mouse ES cells.
Figure 5.
Filter performance in mouse ES cells at sensitivity 20%. The best relative FP rate (as defined in the Methods section) of each filter type has been plotted for the 18 mouse gold-standard TFBS datasets. Multiple gold-standard datasets were available for Klf4, Oct4 and Nanog, and the first author of the corresponding gold-standard dataset has been indicated. PhastCons filtering failed to yield a positive relative FP rate improvement for any of the 18 gold-standard datasets at this sensitivity level, and so has been omitted. Error bars indicate standard error. Barplot mean and standard errors smaller than −1 have been truncated to −1, to allow clearer visualization of relative FP improvement values between 0 and 1.
Figure 6.
Filter performance in mouse ES cells at sensitivity 80%. The best relative FP rate (as defined in the Methods section) of each filter type has been plotted for the TFs cMyc, E2F1, nMyc and Zfx. PhastCons filtering failed to yield a positive relative FP rate improvement for any of the four gold-standard datasets at this sensitivity level, and so has been omitted. Error bars indicate standard error. For a given TF and filter, if the filter cannot attain a sensitivity of 80% due to actual positive elimination, then the bar is omitted from the plot.
Figure 7.
Tissue specificity of TFBS predictions in three human tissues. The best relative FP rate (as defined in the Methods section) of each H3K4me3 filter is shown for the 10 human gold-standard TFBS datasets. Each arrow indicates the results for the H3K4me3 filter using data estimated from the same tissue as the given TFBS gold-standard data. For example, the distribution of HNF4A TFBSs was estimated in liver, so the arrow points to the liver results for HNF4A. Error bars indicate standard error. Barplot mean and standard errors smaller than −1 have been truncated to −1, to allow clearer visualization of relative FP improvement values between 0 and 1.
Figure 8.
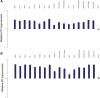
Performance of H3K4me3 filtering without optimization of threshold. The relative FP rate has been plotted for a H3K4me3 filter, with a threshold of 1.0 at a sensitivity of 20% (a) and a more stringent threshold of 2.0 at the lower sensitivity of 10% (b). Error bars indicate standard error. Note that the results presented are relative FP improvement of a filter with a single given threshold, rather than best relative FP improvement. That is, we have not optimized the filtering threshold used.
Figure 9.
Overlap between H3K4me3 and TF occupancy in ES cells at the Bmp4 (a) and Otx2 (b) gene loci. The track labelled ‘ES_K4 wig’ indicates the distribution of H3K4me3 in mouse ES cells, as published by Mikkelsen et al. (5). Units of H3K4me3 density are described in the Methods section. UCSC KnownGenes and NIA Genes are shown in the lowest two tracks for each displayed region. CAGE TU locations are indicated, as are binding locations for TFs Nanog, Oct4, Klf2, Klf4 and Klf5 estimated by Jiang et al. (23) and Loh et al. (31). Red boxes indicate regions at which the available H3K4me3 information should be of greater benefit to TFBS prediction, compared with the available TSS location information, due to the large distance between the TFBSs and known TSSs.
Similar articles
- Histone H3 lysine 9 methylation and HP1gamma are associated with transcription elongation through mammalian chromatin.
Vakoc CR, Mandat SA, Olenchock BA, Blobel GA. Vakoc CR, et al. Mol Cell. 2005 Aug 5;19(3):381-91. doi: 10.1016/j.molcel.2005.06.011. Mol Cell. 2005. PMID: 16061184 - Computer and statistical analysis of transcription factor binding and chromatin modifications by ChIP-seq data in embryonic stem cell.
Orlov Y, Xu H, Afonnikov D, Lim B, Heng JC, Yuan P, Chen M, Yan J, Clarke N, Orlova N, Huss M, Gunbin K, Podkolodnyy N, Ng HH. Orlov Y, et al. J Integr Bioinform. 2012 Sep 18;9(2):211. doi: 10.2390/biecoll-jib-2012-211. J Integr Bioinform. 2012. PMID: 22987856 - Integrating genomic data to predict transcription factor binding.
Holloway DT, Kon M, DeLisi C. Holloway DT, et al. Genome Inform. 2005;16(1):83-94. Genome Inform. 2005. PMID: 16362910 - Transcription factors, chromatin and cancer.
Thorne JL, Campbell MJ, Turner BM. Thorne JL, et al. Int J Biochem Cell Biol. 2009 Jan;41(1):164-75. doi: 10.1016/j.biocel.2008.08.029. Epub 2008 Sep 2. Int J Biochem Cell Biol. 2009. PMID: 18804550 Review. - Transcription factor access to promoter elements.
Morse RH. Morse RH. J Cell Biochem. 2007 Oct 15;102(3):560-70. doi: 10.1002/jcb.21493. J Cell Biochem. 2007. PMID: 17668451 Review.
Cited by
- Genome-wide prediction of transcription factor binding sites using an integrated model.
Won KJ, Ren B, Wang W. Won KJ, et al. Genome Biol. 2010 Jan 22;11(1):R7. doi: 10.1186/gb-2010-11-1-r7. Genome Biol. 2010. PMID: 20096096 Free PMC article. - Sequence and chromatin determinants of cell-type-specific transcription factor binding.
Arvey A, Agius P, Noble WS, Leslie C. Arvey A, et al. Genome Res. 2012 Sep;22(9):1723-34. doi: 10.1101/gr.127712.111. Genome Res. 2012. PMID: 22955984 Free PMC article. - Epigenetic priors for identifying active transcription factor binding sites.
Cuellar-Partida G, Buske FA, McLeay RC, Whitington T, Noble WS, Bailey TL. Cuellar-Partida G, et al. Bioinformatics. 2012 Jan 1;28(1):56-62. doi: 10.1093/bioinformatics/btr614. Epub 2011 Nov 8. Bioinformatics. 2012. PMID: 22072382 Free PMC article. - Assessing phylogenetic motif models for predicting transcription factor binding sites.
Hawkins J, Grant C, Noble WS, Bailey TL. Hawkins J, et al. Bioinformatics. 2009 Jun 15;25(12):i339-47. doi: 10.1093/bioinformatics/btp201. Bioinformatics. 2009. PMID: 19478008 Free PMC article. - A comprehensive review of computational prediction of genome-wide features.
Xu T, Zheng X, Li B, Jin P, Qin Z, Wu H. Xu T, et al. Brief Bioinform. 2020 Jan 17;21(1):120-134. doi: 10.1093/bib/bby110. Brief Bioinform. 2020. PMID: 30462144 Free PMC article.
References
- Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. - PubMed
- Guccione E, Martinato F, Finocchiaro G, Luzi L, Tizzoni L, Dall'O;lio V, Zardo G, Nervi C, Bernard L, Amati B. Myc-binding-site recognition in the human genome is determined by chromatin context. Nat. Cell Biol. 2006;8:764–U225. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous