The structure of F-pili - PubMed (original) (raw)
The structure of F-pili
Ying A Wang et al. J Mol Biol. 2009.
Abstract
Exchange of DNA between bacteria involves conjugative pili. While the prevailing view has been that F-pili are completely retracted before single-stranded DNA is passed from one cell to another, it has recently been reported that the F-pilus, in addition to establishing the contact between mating cells, serves as a channel for passing DNA between spatially separated cells during conjugation. The structure and function of F-pili are poorly understood. They are built from a single subunit having only 70 residues, and the small size of the subunit has made these filaments difficult to study. Here, we have applied electron cryo-microscopy and single-particle methods to solve the long-existing ambiguity in the packing geometry of F-pilin subunits. We show that the F-pilus has an entirely different symmetry from any of the known bacterial pili as well as any of the filamentous bacteriophages, which have been suggested to be structural homologs. Two subunit packing schemes were identified: one has stacked rings of four subunits axially spaced by approximately 12.8 A, while the other has a one-start helical symmetry with an axial rise of approximately 3.5 A per subunit and a pitch of approximately 12.2 A. Both structures have a central lumen of approximately 30 A diameter that is more than large enough to allow for the passage of single-stranded DNA. Remarkably, both schemes appear to coexist within the same filaments, in contrast to filamentous phages that have been described as belonging to one of two possible symmetry classes. For the segments composed of rings, the twist between adjacent rings is quite variable, while the segments having a one-start helix are in multiple states of both twist and extension. This coexistence of two very different symmetries is similar to what has recently been reported for an archaeal Methanococcus maripaludis pili filament and an archaeal Sulfolobus shibatae flagellar filament.
Figures
Figure 1
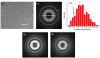
Electron micrograph of frozen-hydrated F-pili (a). The space bar is 1000 Å. F-pili were prepared by a method found to minimize aggregation. F+ cells lacking type I pili were grown to an optical density of 2.0. Cells were rapidly chilled by addition of crushed ice, harvested by sedimentation, and suspended in 3 mL/L of culture of 60% (w/v) sucrose in 20 mM Tris-HCl, pH 7.6. The suspension was brought to 30% sucrose with Tris buffer and cells were removed by sedimentation. F-pili were purified from the supernatant fraction by two cycles of isopycnic sedimentation in sucrose gradients (density range = 1.16 to 1.29 g/mL). Fractions at ∼1.2 g/mL were examined by fluorescence microscopy for F-pili and by SDS gel electrophoresis for purity (where preparations were judged to be >90% F-pilin). Images of frozen-hydrated F-pili were collected on a Tecnai 20 field emission gun microscope at 200 keV. Imaging and analysis were done by similar methods as described previously. Negatives were scanned with a Nikon Coolscan 8000 as 16-bit images using a raster of 2.4 Å/pixel. 10 images with defocus values in the range of 1.5–2.7 μm were selected for further processing, since the n=4 layer line has a maximum near the first maximum of the contrast transfer function (CTF) at these defocus values. All cryo-EM images were multiplied by the theoretical CTF to correct for phase reversals and to optimize the signal-to-noise ratio. An averaged power spectrum (b), generated from 10,952 overlapping segments (each 200 pixels or 480 Å in length) of frozen-hydrated pili shows one layer line (red arrow) at ∼ 1/(32.2 Å) which can be interpreted as either n=4 or n=5, based upon the distance of the peak from the meridian and the diameter of the filaments. Mass per unit length measurements from Scanning Transmission Electron Microscopy (STEM) were made by similar methods as described previously, and yielded an average of 2,368.4 ± 8.0 (SEM) Da/Å (c). Digital STEM dark-field micrographs of freeze-dried specimens were recorded with 512 × 512 pixels at raster steps of 1.0 or 2.0 nm per pixel. The F-pili data were normalized to the known mass-per-unit-length of tobacco mosaic virus (1,314 kDa/nm). Histograms were calculated with 1-kDa/nm bins. A Gaussian was then fit to the distribution using the Origin software package (OriginLab company). The 10,952 segments from the cryo-EM images were sorted into two groups by models with either point group symmetry or one-start helical symmetry. 13% of the segments were sorted to have point group symmetry while the remaining 87% were sorted to have a one-start helix. The mass per unit length from the subset of segments reconstructed with point group symmetry (c, cyan arrow) is larger than that of the subset of segments with one-start helical symmetry (c, black arrow). The averaged power spectrum from the segments sorted as having point group symmetry (d) has a meridional layer line (red arrow) at ∼ 1/(12.6 Å). The averaged power spectrum from the segments sorted to have one-start helical symmetry (e) has a layer line (red arrow) at ∼ 1/(32.2 Å) which can be interpreted as n=4, based upon the distance of the peak from the meridian and the diameter of the filaments.
Figure 2
The 1,464 segments of the first class were sorted into six subgroups by differences in the angular rotation between adjacent rings of subunits. The averaged power spectra from the two most populated subgroups (a,b) are much improved compared to the averaged power spectrum before sorting (Fig. 1d), suggesting reduced heterogeneity in each subgroup. The meridional layer line is fixed while the n=4 layer lines shift as expected in the two subgroups (a,b, red lines). The first subgroup was sorted to have a twist of ∼ -30°. The averaged power spectrum (a) can be interpreted as having an n=0 layer line at ∼ 1/(12.6 Å) and an n=4 layer line at 1/(37.1 Å). The second subgroup was sorted to have a twist of ∼ 34°. The averaged power spectrum (b) can be interpreted as having an n=0 layer line at ∼ 1/(12.6 Å) and n=4 at 1/(33.1 Å). The n=-4 layer line is not visible because it would be at a spacing that is near a minimum in the averaged CTF. The reconstruction of the most populated subgroup (d) has a C4 symmetry with an axial rise of 12.8 Å and a twist of 34.2°. The reconstruction has been divided by the weighted sum of the squared CTF functions and filtered to a resolution of ∼ 14 Å as determined by the 0.5 Fourier Shell Correlation criterion. The power spectrum from the projection of this reconstruction (c) matches the corresponding averaged power spectrum (b). The surface thresholds (d-f) have been chosen to enclose 100% of the expected molecular volume. The main connectivity between subunits is along a left-handed eight-start helix (d, red line) and a right-handed four-start helix (d, cyan line). Two cross-sectional contour plots (e,f) that are spaced 7.2 Å away from each other have similar outer diameters (85 Å) and similar size lumens (30 Å).
Figure 3
The 9,488 segments of the set classified as having a one-start helical symmetry were further sorted into three subgroups by a multi-reference sorting against projections of three different initial reconstructions. The averaged power spectrum from one relatively homogeneous subgroup (containing 23% of this group) (a) is much improved compared to the averaged power spectrum before sorting (Fig. 1e), suggesting reduced heterogeneity in this subgroup. The averaged power spectrum (a) can be interpreted as having an n=4 layer line at ∼ 1/(32.2 Å) (upper red arrow) and n=-7 at ∼ 1/(51.0 Å) (lower red arrow). The reconstruction of this subgroup (c) has an axial rise of 3.5 Å and a twist of 99.6° per subunit. The reconstruction has been divided by the weighted sum of the squared CTF functions and filtered to a resolution of ∼ 13 Å as determined by the 0.5 Fourier Shell Correlation criterion. The power spectrum from the projection of this reconstruction (b) matches the corresponding averaged power spectrum (a). The surface thresholds have been chosen to enclose 108% of the expected molecular volume, since there is a loss of inner connectivity when a 100% threshold is used. We expect that this is due to limited resolution. The main connectivity between subunits is along a left-handed seven-start helix (c, red line) and a right-handed four-start helix (c, cyan line). A cross-sectional contour plot (d) shows similar outer diameter (85 Å) and lumen (30 Å) to that in the point group symmetry set (Fig. 2e,f).
Figure 4
A superposition (a) of the reconstructions with C4 symmetry and one-start helical symmetry in which one subunit in each has been aligned (a, black arrow) shows the similarity of structure for subunits and difference in subunit packing. The yellow surface has C4 symmetry with an axial rise of 12.8 Å and a twist of 34.2° (Fig. 2d), while the cyan surface has a one-start helical symmetry with a twist of 99.6° and an axial rise of 3.5 Å. (Fig. 3c). The aligned subunit in the yellow structure is shown as mesh for clarity. A surface lattice (b,c) shows the arrangement of subunits on the surface of a cylinder, using the convention that the cylindrical surface has been cut open and we are looking at the outside of the surface. Within segments having C4 symmetry (b), the strongest observed connectivity between subunits is along the left-handed eight-start helices (red) and the right-handed four-start helices (cyan). Two helical families are labeled in the helical net for the one-start helix (c). The strongest observed connectivity between subunits within segments having a one-start symmetry is along the right-handed four-start helices (cyan) and the left-handed seven-start helices (red). A unit cell has been colored yellow (b,c) in each, showing that despite the differences in symmetry, the local packing is very similar between the two states.
Similar articles
- The structure of an archaeal pilus.
Wang YA, Yu X, Ng SY, Jarrell KF, Egelman EH. Wang YA, et al. J Mol Biol. 2008 Aug 29;381(2):456-66. doi: 10.1016/j.jmb.2008.06.017. Epub 2008 Jun 12. J Mol Biol. 2008. PMID: 18602118 Free PMC article. - Fluorescence assays for F-pili and their application.
Daehnel K, Harris R, Maddera L, Silverman P. Daehnel K, et al. Microbiology (Reading). 2005 Nov;151(Pt 11):3541-3548. doi: 10.1099/mic.0.28159-0. Microbiology (Reading). 2005. PMID: 16272377 - The flagellar filament structure of the extreme acidothermophile Sulfolobus shibatae B12 suggests that archaeabacterial flagella have a unique and common symmetry and design.
Cohen-Krausz S, Trachtenberg S. Cohen-Krausz S, et al. J Mol Biol. 2008 Jan 25;375(4):1113-24. doi: 10.1016/j.jmb.2007.10.048. Epub 2007 Oct 23. J Mol Biol. 2008. PMID: 18068187 - The archaeabacterial flagellar filament: a bacterial propeller with a pilus-like structure.
Trachtenberg S, Cohen-Krausz S. Trachtenberg S, et al. J Mol Microbiol Biotechnol. 2006;11(3-5):208-20. doi: 10.1159/000094055. J Mol Microbiol Biotechnol. 2006. PMID: 16983196 Review. - Type IV pilin structures: insights on shared architecture, fiber assembly, receptor binding and type II secretion.
Hansen JK, Forest KT. Hansen JK, et al. J Mol Microbiol Biotechnol. 2006;11(3-5):192-207. doi: 10.1159/000094054. J Mol Microbiol Biotechnol. 2006. PMID: 16983195 Review.
Cited by
- Surface organelles assembled by secretion systems of Gram-negative bacteria: diversity in structure and function.
Thanassi DG, Bliska JB, Christie PJ. Thanassi DG, et al. FEMS Microbiol Rev. 2012 Nov;36(6):1046-82. doi: 10.1111/j.1574-6976.2012.00342.x. Epub 2012 May 24. FEMS Microbiol Rev. 2012. PMID: 22545799 Free PMC article. Review. - Plasmid Transfer by Conjugation in Gram-Negative Bacteria: From the Cellular to the Community Level.
Virolle C, Goldlust K, Djermoun S, Bigot S, Lesterlin C. Virolle C, et al. Genes (Basel). 2020 Oct 22;11(11):1239. doi: 10.3390/genes11111239. Genes (Basel). 2020. PMID: 33105635 Free PMC article. Review. - Appendages of the cyanobacterial cell.
Schuergers N, Wilde A. Schuergers N, et al. Life (Basel). 2015 Mar 4;5(1):700-15. doi: 10.3390/life5010700. Life (Basel). 2015. PMID: 25749611 Free PMC article. Review. - Cryo-EM of bacterial pili and archaeal flagellar filaments.
Egelman EH. Egelman EH. Curr Opin Struct Biol. 2017 Oct;46:31-37. doi: 10.1016/j.sbi.2017.05.012. Epub 2017 Jun 10. Curr Opin Struct Biol. 2017. PMID: 28609682 Free PMC article. Review. - Antibiotics as a selective driver for conjugation dynamics.
Lopatkin AJ, Huang S, Smith RP, Srimani JK, Sysoeva TA, Bewick S, Karig DK, You L. Lopatkin AJ, et al. Nat Microbiol. 2016 Apr 11;1(6):16044. doi: 10.1038/nmicrobiol.2016.44. Nat Microbiol. 2016. PMID: 27572835 Free PMC article.
References
- Lanka E, Wilkins BM. DNA processing reactions in bacterial conjugation. Annu Rev Biochem. 1995;64:141–69. - PubMed
- Valentine R, Silverman P, Ippen K, Mobach H. The F-pilus of Escherichia coli. Adv Microb Physiol. 1969;3:1–52.
- Brinton CC., Jr The structure, function, synthesis and genetic control of bacterial pili and a molecular model for DNA and RNA transport in gram negative bacteria. Trans N Y Acad Sci. 1965;27:1003–54. - PubMed
- Lawn AM. Morphological features of the pili associated with Escherichia coli K 12 carrying R factors or the F factor. J Gen Microbiol. 1966;45:377–83. - PubMed
- Silverman PM. Towards a structural biology of bacterial conjugation. Mol Microbiol. 1997;23:423–9. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 EB001567-06/EB/NIBIB NIH HHS/United States
- GM035269/GM/NIGMS NIH HHS/United States
- EB001567/EB/NIBIB NIH HHS/United States
- R01 EB001567/EB/NIBIB NIH HHS/United States
- R01 GM035269/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources