Signal transducer and activator of transcription 4 is required for the transcription factor T-bet to promote T helper 1 cell-fate determination - PubMed (original) (raw)
Signal transducer and activator of transcription 4 is required for the transcription factor T-bet to promote T helper 1 cell-fate determination
Vivian T Thieu et al. Immunity. 2008.
Abstract
Transcriptional regulatory networks direct the development of specialized cell types. The transcription factors signal tranducer and activator of transcription 4 (Stat4) and T-bet are required for the interleukin-12 (IL-12)-stimulated development of T helper 1 (Th1) cells, although the hierarchy of activity by these factors has not been clearly defined. In this report, we show that these factors did not function in a linear pathway and that each factor played a unique role in programming chromatin architecture for Th1 gene expression, with subsets of genes depending on Stat4, T-bet, or both for expression in Th1 cells. T-bet was not able to transactivate expression of Stat4-dependent genes in the absence of endogenous Stat4 expression. Thus, T-bet requires Stat4 to achieve complete IL-12-dependent Th1 cell-fate determination. These data provide a basis for understanding how transiently activated and lineage-specific transcription factors cooperate in promoting cellular differentiation.
Figures
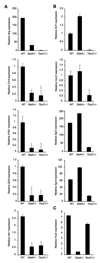
Figure 1. Contribution of Stat4 and T-bet to expression of genes in Th1 cells
Wild type, Stat4-deficient (_Stat4_−/−) and T-bet-deficient (_Tbx21_−/−) CD4+ T cells were cultured under Th1 conditions (IL−12 + anti-IL-4) for five days. RNA was isolated from cells either before (Ccr5, Il18r1, Etv5, Cxcr3) or six hours after (Ifng, Hlx1, Xcl1, Egr2, Egr3, Furin) re-stimulation of cells with anti-CD3. Quantitative PCR using TaqMan primers specific for each gene was performed and results were normalized to expression of beta2-microglobulin. Results are the average ± SD of replicate samples and are representative of four experiments with similar results.
Figure 2. IFNγ or IL-27 do not rescue gene expression in Stat4-deficient Th1 cells
(A) Wild type and Stat4-deficient (_Stat4_−/−) CD4+ T cells were cultured under Th1 conditions (IL-12 + anti-IL-4) in the presence or absence of 100 ng/ml recombinant IFNγ for five days. Cells were re-stimulated with anti-CD3 for 18 hours and supernatants were analyzed for levels of IFNγ and IL-4 using ELISA. (B) Cells cultured as in (A) were analyzed for gene expression using qPCR as described in Figure 1. Results in (A) and (B) the average ± SD of replicate samples and are representative of four experiments with similar results. (C) Wild type and Stat4-deficient (_Stat4_−/−) CD4+ T cells were cultured under Th1 conditions (IL−12 + anti-IL-4) in the presence or absence of 100 ng/ml recombinant IL-27 for five days. Expression of genes was determined after activation with anti-CD3 for four hours. Results are representative of two experiments with similar results.
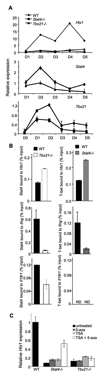
Figure 3. Stat4 and T-bet bind to the Hlx1 locus
(A) Wild type, Stat4-deficient (_Stat4_−/−) and T-bet-deficient (_Tbx21_−/−) CD4+ T cells were cultured under Th1 conditions (IL-12 + anti-IL-4) for five days and RNA was isolated from cells during each day of culture. Expression of Hlx1, Stat4 and Tbx21 were assessed in each of the samples using qPCR. Results are representative of two experiments. (B) Wild type, Stat4-deficient (_Stat4_−/−) and T-bet-deficient (_Tbx21_−/−) CD4+ T cells were cultured under Th1 conditions (IL-12 + anti-IL-4) for five days and chromatin was isolated for ChIP assay. ChIP was performed for Stat4 bound to the promoter of Hlx1, Ifng or Il18r1 in wild type and T-bet-deficient cells (left) or for T-bet bound to the same regions in wild type or Stat4-deficient cells (right). QPCR was performed using TaqMan primers specific for each promoter. Transcription factor bound to the locus is expressed as the percent of the input used for the ChIP assay. Results are the average ± SD of replicate samples and are representative of three experiments for Hlx1 and two experiments for binding to other promoters with similar results. ND, not detected. (C) Wild type, Stat4-deficient (_Stat4_−/−) and T-bet-deficient (_Tbx21_−/−) CD4+ T cells were cultured under Th1 conditions (IL-12 + anti-IL-4) for five days in the presence or absence of 20 nM trichostatin A (TSA) and/or 10 µM 5-aza-deoxycytidine (5-aza). RNA was isolated for analysis of Hlx1 gene expression as described in Figure 1. Results are representative of two experiments.
Figure 4. Stat4- and T-bet-dependent chromatin remodeling at the Hlx1 locus
(A-E) Wild type, Stat4-deficient (_Stat4_−/−) and T-bet-deficient (_Tbx21_−/−) CD4+ T cells were cultured under Th1 conditions (IL-12 + anti-IL-4) for five days and chromatin was isolated for ChIP assay. ChIP was performed for acetylated-H3, -H4 and DNMT3a on days 3–5 of culture (A) or day 5 only (B), the histone acetyltransferases CBP, p300, PCAF and Gcn5 on day 5 of culture (C), acetylated H4K5 and K8 on day 5 of culture (D), or EZH2, H3K27me3, H4K20me3 and H3K4me2 on day 5 of culture (E) using qPCR primers for the Hlx1 promoter. Results are the average ± SD of replicate samples and are representative of 3–5 experiments for each modification or enzyme with similar patterns. ND, not detected.
Figure 5. Stat4- and T-bet-dependent chromatin remodeling at target loci
(A-F) Wild type, Stat4-deficient (_Stat4_−/−) and T-bet-deficient (_Tbx21_−/−) CD4+ T cells were cultured under Th1 conditions (IL-12 + anti-IL-4) for five days and chromatin was isolated for ChIP assay. ChIP assay was performed for acetylated-H3, -H4 and H3K4me2 at the Ifng promoter (−0.4 kb), and at sites +20 kb and +40 kb from the transcriptional start site (A), and at the Furin promoter (B). ChIP assay was performed for DNMT3a (C) and H4K20me3 (D) at the Ifng and Furin promoters. ChIP assay was performed for acetylated-H3, -H4 and H3K4me2 at the Xcl1 promoter (E) and for acetylated-H4 and H3K4me2 at intron 1 of Cd4 (F). Results are the average ± SD of replicate samples and are representative of 2–4 experiments for each modification or enzyme with similar patterns.
Figure 6. Stat4 requirement in T-bet function
(A) Wild type, Stat4-deficient (_Stat4_−/−), T-bet-deficient (_Tbx21_−/−) and Stat4-T-bet-double deficient (_Stat4-Tbx21_−/−) CD4+ T cells were cultured under Th1 conditions (IL-12 + anti-IL-4) for five days and total cell extracts were immunoblotted for T-bet, Stat4 and GAPDH as a control. (B) Cells cultured as in (A) were assessed for the expression of Th1 genes before (Il18rap, Runx3) or after (Lta) re-stimulation with anti-CD3. (C-G) Wild type, T-bet-deficient (_Tbx21_−/−) and Stat4-T-bet-double deficient (_Stat4_−/− _Tbx21_−/−) CD4+ T cells were cultured under Th1 conditions. On day 2 of the culture period, cells were transduced with a bicistronic retrovirus expressing EGFP only (MIEG) or T-bet and EGFP (T-bet). At the end of the culture, cells were sorted for EGFP expression and stimulated for 18 hours with anti-CD3. Supernatants were analyzed for IFNγ levels using ELISA (C). RNA was isolated from each population to determine the expression levels of the indicated genes using qPCR (D). Surface expression of CXCR3 was determined using flow cytometry (E). ChIP assay was performed for acetylated-H3 or -H4 at the Hlx1 and Ifng promoters (F) or acetylated-H4 at the Xcl1 promoter (G). Results are the average ± SD of replicate samples and are representative of 2–3 experiments with similar results.
Similar articles
- Stat4 is critical for the balance between Th17 cells and regulatory T cells in colitis.
Xu J, Yang Y, Qiu G, Lal G, Yin N, Wu Z, Bromberg JS, Ding Y. Xu J, et al. J Immunol. 2011 Jun 1;186(11):6597-606. doi: 10.4049/jimmunol.1004074. Epub 2011 Apr 27. J Immunol. 2011. PMID: 21525389 Free PMC article. - [Impact of cyclosporine A on the expression of T-bet, GATA-3, relevant signal transduction molecules, cytokine and Th1/Th2 balance in patients with chronic aplastic anemia].
Li J, Zhou YM, Hu MH, Sun WL, Xue ZZ. Li J, et al. Zhongguo Shi Yan Xue Ye Xue Za Zhi. 2010 Oct;18(5):1211-9. Zhongguo Shi Yan Xue Ye Xue Za Zhi. 2010. PMID: 21129263 Chinese. - IFN-alpha is not sufficient to drive Th1 development due to lack of stable T-bet expression.
Ramos HJ, Davis AM, George TC, Farrar JD. Ramos HJ, et al. J Immunol. 2007 Sep 15;179(6):3792-803. doi: 10.4049/jimmunol.179.6.3792. J Immunol. 2007. PMID: 17785816 Free PMC article. - Transcription Factor T-bet Orchestrates Lineage Development and Function in the Immune System.
Kallies A, Good-Jacobson KL. Kallies A, et al. Trends Immunol. 2017 Apr;38(4):287-297. doi: 10.1016/j.it.2017.02.003. Epub 2017 Mar 7. Trends Immunol. 2017. PMID: 28279590 Review. - A unique role for IL-23 in promoting cellular immunity.
Lankford CS, Frucht DM. Lankford CS, et al. J Leukoc Biol. 2003 Jan;73(1):49-56. doi: 10.1189/jlb.0602326. J Leukoc Biol. 2003. PMID: 12525561 Review.
Cited by
- STAT4 Directs a Protective Innate Lymphoid Cell Response to Gastrointestinal Infection.
Dulson SJ, Watkins EE, Crossman DK, Harrington LE. Dulson SJ, et al. J Immunol. 2019 Nov 1;203(9):2472-2484. doi: 10.4049/jimmunol.1900719. Epub 2019 Sep 27. J Immunol. 2019. PMID: 31562212 Free PMC article. - Differentiation of effector CD4 T cell populations (*).
Zhu J, Yamane H, Paul WE. Zhu J, et al. Annu Rev Immunol. 2010;28:445-89. doi: 10.1146/annurev-immunol-030409-101212. Annu Rev Immunol. 2010. PMID: 20192806 Free PMC article. Review. - T-bet: a bridge between innate and adaptive immunity.
Lazarevic V, Glimcher LH, Lord GM. Lazarevic V, et al. Nat Rev Immunol. 2013 Nov;13(11):777-89. doi: 10.1038/nri3536. Epub 2013 Oct 11. Nat Rev Immunol. 2013. PMID: 24113868 Free PMC article. Review. - Enteric pathogens and gut function: Role of cytokines and STATs.
Shea-Donohue T, Fasano A, Smith A, Zhao A. Shea-Donohue T, et al. Gut Microbes. 2010 Sep;1(5):316-324. doi: 10.4161/gmic.1.5.13329. Epub 2010 May 12. Gut Microbes. 2010. PMID: 21327040 Free PMC article. - Mechanisms of Antiviral Cytotoxic CD4 T Cell Differentiation.
Knudson CJ, Férez M, Alves-Peixoto P, Erkes DA, Melo-Silva CR, Tang L, Snyder CM, Sigal LJ. Knudson CJ, et al. J Virol. 2021 Sep 9;95(19):e0056621. doi: 10.1128/JVI.00566-21. Epub 2021 Jul 14. J Virol. 2021. PMID: 34260270 Free PMC article.
References
- Afkarian M, Sedy JR, Yang J, Jacobson NG, Cereb N, Yang SY, Murphy TL, Murphy KM. T-bet is a STAT1-induced regulator of IL-12R expression in naive CD4+ T cells. Nat Immunol. 2002;3:549–557. - PubMed
- Agalioti T, Chen G, Thanos D. Deciphering the transcriptional histone acetylation code for a human gene. Cell. 2002;111:381–392. - PubMed
- Ansel KM, Lee DU, Rao A. An epigenetic view of helper T cell differentiation. Nat Immunol. 2003;4:616–623. - PubMed
- Avni O, Lee D, Macian F, Szabo SJ, Glimcher LH, Rao A. T(H) cell differentiation is accompanied by dynamic changes in histone acetylation of cytokine genes. Nat Immunol. 2002;3:643–651. - PubMed
- Chang HC, Zhang S, Thieu VT, Slee RB, Bruns HA, Laribee RN, Klemsz MJ, Kaplan MH. PU.1 expression delineates heterogeneity in primary Th2 cells. Immunity. 2005;22:693–703. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R56 AI045515/AI/NIAID NIH HHS/United States
- R01 AI045515-09/AI/NIAID NIH HHS/United States
- AI45515/AI/NIAID NIH HHS/United States
- AI57459/AI/NIAID NIH HHS/United States
- R01 AI057459-03/AI/NIAID NIH HHS/United States
- R01 AI057459/AI/NIAID NIH HHS/United States
- R01 AI045515/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous