Crystal structure of FadA adhesin from Fusobacterium nucleatum reveals a novel oligomerization motif, the leucine chain - PubMed (original) (raw)
Crystal structure of FadA adhesin from Fusobacterium nucleatum reveals a novel oligomerization motif, the leucine chain
Stanley Nithianantham et al. J Biol Chem. 2009.
Abstract
Many bacterial appendages have filamentous structures, often composed of repeating monomers assembled in a head-to-tail manner. The mechanisms of such linkages vary. We report here a novel protein oligomerization motif identified in the FadA adhesin from the Gram-negative bacterium Fusobacterium nucleatum. The 2.0 angstroms crystal structure of the secreted form of FadA (mFadA) reveals two antiparallel alpha-helices connected by an intervening 8-residue hairpin loop. Leucine-leucine contacts play a prominent dual intra- and intermolecular role in the structure and function of FadA. First, they comprise the main association between the two helical arms of the monomer; second, they mediate the head-to-tail association of monomers to form the elongated polymers. This leucine-mediated filamentous assembly of FadA molecules constitutes a novel structural motif termed the "leucine chain." The essential role of these residues in FadA is corroborated by mutagenesis of selected leucine residues, which leads to the abrogation of oligomerization, filament formation, and binding to host cells.
Figures
FIGURE 1.
Amino acid sequence of FadA in one-letter code. The_underlined_ residues encode the signal peptide followed by mFadA. The leucine residues are marked by asterisks, the central hairpin residues TRFY are marked by dotted lines, and the histidine tag with the extra leucine and glutamic acid residues at the C terminus are_italicized in gray_. In all assays, the His-tagged proteins were used.
FIGURE 2.
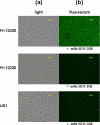
Immunofluorescent staining of wild-type F. nucleatum 12230 and the fadA deletion mutant, US1, using anti-FadA mAb 5G11-3G8 as primary antibodies and FITC-conjugated goat-anti-mouse IgG as secondary antibodies. F. nucleatum 12230 was also incubated directly with the secondary antibodies without prior incubation with mAb 5G11-3G8. The same fields were observed under both light (a) and fluorescence (b) microscope using a ×40 objective.
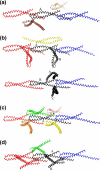
FIGURE 3.
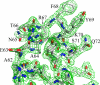
Crystal structure of mFadA. a, ribbon diagram of mFadA. Leucine residues are shown in ball-and-stick representation. b, leucine and tyrosine residues in the vicinity of the hairpin loop. The hydroxyl group of tyrosine is shown in red. c, a trimer of mFadA formed by the interaction of the monomer in the asymmetric unit with neighboring molecules related by a simple translation along the crystallographic b axis. Leucines and residues of the hairpin loop are shown in ball-and-stick representation.
FIGURE 4.
2_Fo_-Fc electron density map of the hairpin region at 2. 0 Å resolution at contour level of 1.4 standard deviations above the mean. mFadA is shown in stick representation. All labeled residues were omitted from the phase calculation.
FIGURE 5.
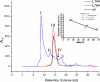
FPLC Superdex S-200 size exclusion chromatography of wild-type and variant mFadA. _A_280 represents the absorbance at 280 nm in arbitrary units. Wild-type (wt) mFadA eluted as five major peaks I, II, III, IV, and V, possibly corresponding to polymer, tetramer, trimer, dimer, and monomer, respectively. The L14A and L76A variant mFadA did not form aggregates higher than tetramer. Inset, molecular weight (MW) calibration curve with the proteins lysozyme, TolB, and bovine serum albumin (BSA).
FIGURE 6.
Variation of interfaces in wild-type and variant proteins. Molecules are shown as Cα traces. The central molecule is shown in black. a, the central molecule of wild-type mFadA and four neighboring molecules. b, L14A form I, with molecule A (top) and molecule B (bottom) in the asymmetric unit. c, L14A form II.d, L76A. The head-to-tail interactions are conserved in all the structures, whereas other intermolecular interactions vary.
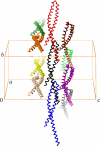
FIGURE 7.
Packing diagram of the wild-type mFadA crystal structure. Individual protomers are shown in α-carbon representation in different colors. The unit cell axes a, b, and c are indicated. Note that the helical axes are roughly parallel to the crystallographic b axis.
FIGURE 8.
Electron micrographs of FadA. a, wild type; b, L14A; and c, L76A. Magnification: ×50,000. wt, wild type.
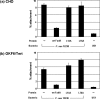
FIGURE 9.
Inhibition of attachment of F. nucleatum 12230 (F. nuc 12230) to Chinese hamster ovary (CHO)(a) and OKF6/Tert (b) cells by wild-type FadA (wt FadA) and its variants. F. nucleatum 12230 was added to the monolayers either directly or following preincubation of wild-type FadA (50 μg/well), L14A, or L76A variants (each at 200 μg/well). F. nucleatum 12230 US1 was added directly to the host cells as a negative control.
References
- Kang, H. J., Coulibaly, F., Clow, F., Proft, T., and Baker, E. N. (2007) Science 318 1625-1628 - PubMed
- Moore, W. E., and Moore, L. V. (1994) Periodontol. 2000 5 66-77 - PubMed
- Chaim, W., and Mazor, M. (1992) Arch. Gynecol. Obstet. 251 9-15 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources