INTERFEROME: the database of interferon regulated genes - PubMed (original) (raw)
. 2009 Jan;37(Database issue):D852-7.
doi: 10.1093/nar/gkn732. Epub 2008 Nov 7.
Affiliations
- PMID: 18996892
- PMCID: PMC2686605
- DOI: 10.1093/nar/gkn732
INTERFEROME: the database of interferon regulated genes
Shamith A Samarajiwa et al. Nucleic Acids Res. 2009 Jan.
Abstract
INTERFEROME is an open access database of types I, II and III Interferon regulated genes (http://www.interferome.org) collected from analysing expression data sets of cells treated with IFNs. This database of interferon regulated genes integrates information from high-throughput experiments with annotation, ontology, orthologue sequences from 37 species, tissue expression patterns and gene regulatory information to enable a detailed investigation of the molecular mechanisms underlying IFN biology. INTERFEROME fulfils a need in infection, immunity, development and cancer research by providing computational tools to assist in identifying interferon signatures in gene lists generated by high-throughput expression technologies, and their potential molecular and biological consequences.
Figures
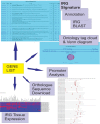
Figure 1.
The functionality of the INTERFEROME database is summarized in Figure 1, which displays some of the screen captures resulting from searching the database with a gene list. Its main functionality is the IRG signature page which identifies IRGs that were present in the gene list and displays associated annotation. In addition INTERFEROME's expression and promoter analysis capability is highlighted. An example search is demonstrated in
Supplementary Figures
.
Similar articles
- Interferome v2.0: an updated database of annotated interferon-regulated genes.
Rusinova I, Forster S, Yu S, Kannan A, Masse M, Cumming H, Chapman R, Hertzog PJ. Rusinova I, et al. Nucleic Acids Res. 2013 Jan;41(Database issue):D1040-6. doi: 10.1093/nar/gks1215. Epub 2012 Nov 29. Nucleic Acids Res. 2013. PMID: 23203888 Free PMC article. - Systems biology of interferon responses.
Hertzog P, Forster S, Samarajiwa S. Hertzog P, et al. J Interferon Cytokine Res. 2011 Jan;31(1):5-11. doi: 10.1089/jir.2010.0126. J Interferon Cytokine Res. 2011. PMID: 21226606 Review. - Functional classification of interferon-stimulated genes identified using microarrays.
de Veer MJ, Holko M, Frevel M, Walker E, Der S, Paranjape JM, Silverman RH, Williams BR. de Veer MJ, et al. J Leukoc Biol. 2001 Jun;69(6):912-20. J Leukoc Biol. 2001. PMID: 11404376 Review. - Tissue and time specific expression pattern of interferon regulated genes in the chicken.
Röll S, Härtle S, Lütteke T, Kaspers B, Härtle S. Röll S, et al. BMC Genomics. 2017 Mar 28;18(1):264. doi: 10.1186/s12864-017-3641-6. BMC Genomics. 2017. PMID: 28351377 Free PMC article. - Interferome signature dynamics during the anti-dengue immune response: a systems biology characterization.
Usuda JN, Plaça DR, Fonseca DLM, Marques AHC, Filgueiras IS, Chaves VGB, Adri AS, Torrentes-Carvalho A, Hirata MH, Freire PP, Catar R, Cabral-Miranda G, Schimke LF, Moll G, Cabral-Marques O. Usuda JN, et al. Front Immunol. 2023 Aug 10;14:1243516. doi: 10.3389/fimmu.2023.1243516. eCollection 2023. Front Immunol. 2023. PMID: 37638052 Free PMC article.
Cited by
- Interferome v2.0: an updated database of annotated interferon-regulated genes.
Rusinova I, Forster S, Yu S, Kannan A, Masse M, Cumming H, Chapman R, Hertzog PJ. Rusinova I, et al. Nucleic Acids Res. 2013 Jan;41(Database issue):D1040-6. doi: 10.1093/nar/gks1215. Epub 2012 Nov 29. Nucleic Acids Res. 2013. PMID: 23203888 Free PMC article. - Structural and dynamic determinants of type I interferon receptor assembly and their functional interpretation.
Piehler J, Thomas C, Garcia KC, Schreiber G. Piehler J, et al. Immunol Rev. 2012 Nov;250(1):317-34. doi: 10.1111/imr.12001. Immunol Rev. 2012. PMID: 23046138 Free PMC article. Review. - Permissive and restricted virus infection of murine embryonic stem cells.
Wash R, Calabressi S, Franz S, Griffiths SJ, Goulding D, Tan EP, Wise H, Digard P, Haas J, Efstathiou S, Kellam P. Wash R, et al. J Gen Virol. 2012 Oct;93(Pt 10):2118-2130. doi: 10.1099/vir.0.043406-0. Epub 2012 Jul 18. J Gen Virol. 2012. PMID: 22815272 Free PMC article. - IL7Rα expression and upregulation by IFNβ in dendritic cell subsets is haplotype-dependent.
McKay FC, Hoe E, Parnell G, Gatt P, Schibeci SD, Stewart GJ, Booth DR. McKay FC, et al. PLoS One. 2013 Oct 16;8(10):e77508. doi: 10.1371/journal.pone.0077508. eCollection 2013. PLoS One. 2013. PMID: 24147013 Free PMC article. - Human cytomegalovirus IE1 protein elicits a type II interferon-like host cell response that depends on activated STAT1 but not interferon-γ.
Knoblach T, Grandel B, Seiler J, Nevels M, Paulus C. Knoblach T, et al. PLoS Pathog. 2011 Apr;7(4):e1002016. doi: 10.1371/journal.ppat.1002016. Epub 2011 Apr 14. PLoS Pathog. 2011. PMID: 21533215 Free PMC article.
References
- Stark GR, Kerr IM, Williams BR, Silverman RH, Schreiber RD. How cells respond to interferons. Annu. Rev. Biochem. 1998;67:227–264. - PubMed
- Hertzog PJ, O’Neill LA, Hamilton JA. The interferon in TLR signaling: more than just antiviral. Trends Immunol. 2003;24:534–539. - PubMed
- Pestka S, Langer JA, Zoon KC, Samuel CE. Interferons and their actions. Annu. Rev. Biochem. 1987;56:727–777. - PubMed
- Schmidt KN, Ouyang W. Targeting interferon-alpha: a promising approach for systemic lupus erythematosus therapy. Lupus. 2004;13:348–352. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources