Role for perinuclear chromosome tethering in maintenance of genome stability - PubMed (original) (raw)
Role for perinuclear chromosome tethering in maintenance of genome stability
Karim Mekhail et al. Nature. 2008.
Abstract
Repetitive DNA sequences, which constitute half the genome in some organisms, often undergo homologous recombination. This can instigate genomic instability resulting from a gain or loss of DNA. Assembly of DNA into silent chromatin is generally thought to serve as a mechanism ensuring repeat stability by limiting access to the recombination machinery. Consistent with this notion is the observation, in the budding yeast Saccharomyces cerevisiae, that stability of the highly repetitive ribosomal DNA (rDNA) sequences requires a Sir2-containing chromatin silencing complex that also inhibits transcription from foreign promoters and transposons inserted within the repeats by a process called rDNA silencing. Here we describe a protein network that stabilizes rDNA repeats of budding yeast by means of interactions between rDNA-associated silencing proteins and two proteins of the inner nuclear membrane (INM). Deletion of either the INM or silencing proteins reduces perinuclear rDNA positioning, disrupts the nucleolus-nucleoplasm boundary, induces the formation of recombination foci, and destabilizes the repeats. In addition, artificial targeting of rDNA repeats to the INM suppresses the instability observed in cells lacking an rDNA-associated silencing protein that is typically required for peripheral tethering of the repeats. Moreover, in contrast to Sir2 and its associated nucleolar factors, the INM proteins are not required for rDNA silencing, indicating that Sir2-dependent silencing is not sufficient to inhibit recombination within the rDNA locus. These findings demonstrate a role for INM proteins in the perinuclear localization of chromosomes and show that tethering to the nuclear periphery is required for the stability of rDNA repeats. The INM proteins studied here are conserved and have been implicated in chromosome organization in metazoans. Our results therefore reveal an ancient mechanism in which interactions between INM proteins and chromosomal proteins ensure genome stability.
Figures
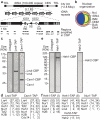
Figure 1. Protein network extending from rDNA to the nuclear envelope
a, rDNA repeats on Chr. XII. Each unit yields a Pol I-transcribed 35S precursor rRNA (processed into 25S, 18S, and 5.8S) and a Pol III-transcribed 5S rRNA. CEN, centromere; TEL, telomere; IGS, intergenic spacer; RE, recombination enhancer;, replication fork block; TIR, Pol I transcription initiation region; O, DNA replication origin; Mbp, megabase pairs. Vertical arrowheads indicate insertion sites of mURA3 reporter genes used in this study. b, Nuclear organization at the G2/M cell cycle stage. No1., nucleolus; ONM, outer nuclear membrane. c-f, Purification of native complexes. c, e, Protein detection in silver-stained gels. TAP cleavage during protein purification leaves a calmodulin binding protein (CBP) fragment. d, f, LC-MS/MS analysis. Number of unique peptides followed by percent coverage of the protein sequence is shown. SI contains full protein lists (Supplementary Table 1, part A) and spectral counts (Supplementary Tables 2 and 3).
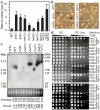
Figure 2. Role of perinuclear protein network at rDNA repeats
a, b, Rates of ADE2 marker loss (mean ± s.d.) relative to wild-type (WT) (a) and representative colonies (b) are shown. c, CHEF analysis of rDNA stability. Chromosomes resolved by CHEF were probed with IGS1 rDNA (Chr. XII, top). Ethidium bromide (EtBr) staining of Chr. IV and smaller chromosomes shows quality of the preparation (bottom). Values corresponding to rDNA copy-number averages and smear edges are indicated. Chr. XII size for sir2Δ was too heterogeneous to estimate copy-number. d, Unlike Sir2 and Cohibin, CLIP is dispensable for rDNA silencing. Ten-fold serial dilutions of cells with the mURA3 reporter gene inserted at IGS1/IGS2 (see Fig. 1a for locations) or outside rDNA at the LEU2 locus are shown.
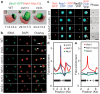
Figure 3. Protein network tethers rDNA to the nuclear envelope
a, Representative 3D reconstructions of cells from double immunofluorescence analysis reveal the relative organization of Net1-GFP (green) and Heh1-Myc13 (red). Signal boundaries are shown with clipping planes for Heh1-Myc13. Percentages of green relative to full red volumes are shown (mean ± s.d.; n = 5). b, Nocodazole-arrested (G2/M) cells were subjected to FISH to visualize rDNA and DAPI staining (pseudocoloured red) to visualize bulk nuclear DNA. Cells with different rDNA morphologies are indicated by double arrowheads (spooled lines), arrows (amorphous), or triangles (two bodies) with quantifications in Supplementary Fig. 6b. Scale bar, 3 μm. c, Live cells with TetI-RFP-marked rDNA and expressing Rad52-YFP and nucleolar Nop1-CFP were imaged. Images depicting most observed phenotypes are shown. More images and quantifications are in Supplementary Fig. 6d-f. Scale bars are 1 μm (white) and 3 μm (black). d, e, Relative fold enrichment of indicated TAP-tagged proteins are shown. rDNA organization schematics are shown on graphs. Gels for d and e are shown in Supplementary Figs 7a and 7d, respectively. Detailed IGS1 ChIP is shown in Supplementary Fig. 7b, c.
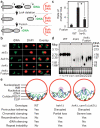
Figure 4. Targeted perinuclear tethering promotes rDNA repeat stability
a, Fusion of Heh1 to Sir2 in lrs4Δ cells. Heh1 is shown embedded in the INM. b, FISH reveals that fusion of Heh1 and Sir2 restores the separation of rDNA signal from DAPI staining in nocodazole-arrested cells lacking Lrs4. Scale bar, 3 μm. c, Relative rates of ADE2 marker loss (mean ± s.d.). Asterisk, P < 0.0001 for Student's t test. d, CHEF analysis of rDNA stability. Chromosomes resolved by CHEF were probed with IGS1 rDNA (Chr. XII, top). EtBr staining of Chr. IV and smaller chromosomes shows quality of the preparation (bottom). e, Functional organization of the perinuclear molecular network tethering rDNA to the nuclear periphery. Repeat instability results from the loss of either the INM CLIP proteins or rDNA silencing complexes.
Similar articles
- Linker histone H1 represses recombination at the ribosomal DNA locus in the budding yeast Saccharomyces cerevisiae.
Li C, Mueller JE, Elfline M, Bryk M. Li C, et al. Mol Microbiol. 2008 Feb;67(4):906-19. doi: 10.1111/j.1365-2958.2007.06101.x. Epub 2008 Jan 7. Mol Microbiol. 2008. PMID: 18179596 - Yeast histone H3 lysine 4 demethylase Jhd2 regulates mitotic rDNA condensation.
Ryu HY, Ahn S. Ryu HY, et al. BMC Biol. 2014 Sep 24;12:75. doi: 10.1186/s12915-014-0075-3. BMC Biol. 2014. PMID: 25248920 Free PMC article. - [Recombination regulation by noncoding transcription in yeast rDNA repeats].
Kobayashi T. Kobayashi T. Tanpakushitsu Kakusan Koso. 2006 Nov;51(14 Suppl):2141-3. Tanpakushitsu Kakusan Koso. 2006. PMID: 17471925 Review. Japanese. No abstract available. - DNA protein interactions at the rRNA of Saccharomyces cerevisiae.
Cioci F, Di Felice F, Chiani F, Camilloni G. Cioci F, et al. Ital J Biochem. 2007 Jun;56(2):81-90. Ital J Biochem. 2007. PMID: 17722648 Review.
Cited by
- RNA-cDNA hybrids mediate transposition via different mechanisms.
Todd LA, Hall AC, Pietrobon V, Chan JNY, Laflamme G, Mekhail K. Todd LA, et al. Sci Rep. 2020 Sep 29;10(1):16034. doi: 10.1038/s41598-020-73018-y. Sci Rep. 2020. PMID: 32994470 Free PMC article. - Intersection of calorie restriction and magnesium in the suppression of genome-destabilizing RNA-DNA hybrids.
Abraham KJ, Chan JN, Salvi JS, Ho B, Hall A, Vidya E, Guo R, Killackey SA, Liu N, Lee JE, Brown GW, Mekhail K. Abraham KJ, et al. Nucleic Acids Res. 2016 Oct 14;44(18):8870-8884. doi: 10.1093/nar/gkw752. Epub 2016 Aug 29. Nucleic Acids Res. 2016. PMID: 27574117 Free PMC article. - Nucleophagy-Implications for Microautophagy and Health.
Bo Otto F, Thumm M. Bo Otto F, et al. Int J Mol Sci. 2020 Jun 24;21(12):4506. doi: 10.3390/ijms21124506. Int J Mol Sci. 2020. PMID: 32599961 Free PMC article. Review. - DNA replication and replication stress response in the context of nuclear architecture.
González-Acosta D, Lopes M. González-Acosta D, et al. Chromosoma. 2024 Jan;133(1):57-75. doi: 10.1007/s00412-023-00813-7. Epub 2023 Dec 6. Chromosoma. 2024. PMID: 38055079 Free PMC article. Review. - Native Chromatin Proteomics Reveals a Role for Specific Nucleoporins in Heterochromatin Organization and Maintenance.
Iglesias N, Paulo JA, Tatarakis A, Wang X, Edwards AL, Bhanu NV, Garcia BA, Haas W, Gygi SP, Moazed D. Iglesias N, et al. Mol Cell. 2020 Jan 2;77(1):51-66.e8. doi: 10.1016/j.molcel.2019.10.018. Epub 2019 Nov 26. Mol Cell. 2020. PMID: 31784357 Free PMC article.
References
- Moazed D, Johnson D. A deubiquitinating enzyme interacts with SIR4 and regulates silencing in S. cerevisiae. Cell. 1996;86:667–677. - PubMed
- Haas W, et al. Optimization and use of peptide mass measurement accuracy in shotgun proteomics. Mol. Cell. Proteomics. 2006;5:1326–1337. - PubMed
- Buker SM, et al. Two different Argonaute complexes are required for siRNA generation and heterochromatin assembly in fission yeast. Nat. Struct. Mol. Biol. 2007;14:200–207. - PubMed
- Beausoleil SA, Villen J, Gerber SA, Rush J, Gygi SP. A probability-based approach for high-throughput protein phosphorylation analysis and site localization. Nat. Biotechnol. 2006;24:1285–1292. - PubMed
- Kurdistani SK, Grunstein M. In vivo protein-protein and protein-DNA crosslinking for genomewide binding microarray. Methods. 2003;31:90–95. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases