Genome-wide polysome profiling reveals an inflammation-responsive posttranscriptional operon in gamma interferon-activated monocytes - PubMed (original) (raw)
Genome-wide polysome profiling reveals an inflammation-responsive posttranscriptional operon in gamma interferon-activated monocytes
Keyur Vyas et al. Mol Cell Biol. 2009 Jan.
Abstract
We previously showed that ribosomal protein L13a is required for translational silencing of gamma interferon (IFN-gamma)-induced ceruloplasmin (Cp) synthesis in monocytes. This silencing also requires the presence of the GAIT (IFN-gamma activated inhibitor of translation) element in the 3' untranslated region (UTR) of Cp mRNA. Considering that Cp is an inflammatory protein, we hypothesized that this mechanism may have evolved to silence a family of proinflammatory proteins, of which Cp is just one member. To identify the other mRNAs that are targets for this silencing, we performed a genome-wide analysis of the polysome-profiled mRNAs by using an Affymetrix GeneChip and an inflammation-responsive gene array. A cluster of mRNAs encoding different chemokines and their receptors was identified as common hits in the two approaches and validated by real-time PCR. In silico predicted GAIT hairpins in the 3' UTRs of the target mRNAs were confirmed as functional cis-acting elements for translational silencing by luciferase reporter assays. Consistent with Cp, the newly identified target mRNAs also required L13a for silencing. Our studies have identified a new inflammation-responsive posttranscriptional operon that can be regulated directly at the level of translation in IFN-gamma-activated monocytes. This regulation of a cohort of mRNAs encoding inflammatory proteins may be important to resolve inflammation.
Figures
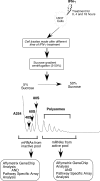
FIG. 1.
Flow chart of the isolation of translationally active and inactive mRNAs from IFN-γ treated U937 cells for different time.
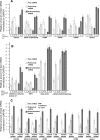
FIG. 2.
Heat map generated from the GeneChip analysis of translationally active and inactive pools of mRNAs. Raw expression data were exported from GCOS (Affymetrix, Santa Clara, CA) to the Data Mining Tools program (Affymetrix, Santa Clara, CA) and subjected to SOM cluster analysis to identify groups of genes with expression patterns similar to the Cp pattern, i.e., induction in response to IFN-γ, greater induction in polysome fractions and less in nonpolysome fractions in 4 h, and the reverse at 18 h after IFN-γ treatment. The ratios of the raw signal intensity values of the polysomal to nonpolysomal mRNAs of the selected genes were determined. Heat maps were generated using gene expression data for selected genes using the programs Cluster, version 2.11 (6), and TreeView, version 1.6 (6). For the unfractionated RNA, the raw signal intensity values of the total mRNA obtained from different times of IFN-γ treatment were used. The color progression scale represents the relationship between different colors and relative quantities of a particular mRNA. IL-6st, interleukin-6 signal transducer; USP, ubiquitin-specific protease; HVP, herpesvirus papio.
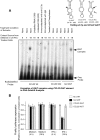
FIG. 3.
Raw data from the mRNA profiling analysis using an inflammatory response pathway array. mRNAs isolated from the translationally active (polysomal) and silenced pools from IFN-γ-treated U937 cells were subjected to microarray analysis using a human inflammatory response microarray (catalog item OHS-803; SuperArray Inc., Frederick, MD). The raw image was generated after chemiluminescence detection, scanned, and analyzed using the GE Expression Analysis Suite, version 2.0 (
http://geasuite.superarray.com
). The results of this analysis are presented in Table S2 in the supplemental material.
FIG. 4.
(A) Comparison of the translational efficiencies of the target mRNAs in normal and L13a-depleted cells using an inflammatory response pathway array. The ratios of polysomal to nonpolysomal abundance of the common mRNAs (i.e., found positive both in GeneChip and inflammatory response pathway arrays) was determined in L13a-depleted and normal U937 cells after 18 h of IFN-γ treatment. Ratios were retrieved from data in Tables S1 and S2 in the supplemental material. (B) Extent of L13a depletion. Immunoblot analysis using anti-L13a antibody was carried out using the lysate made from the cells expressing shRNA against L13a and normal cells (upper panel). The same blot was reprobed with antiactin antibody as a control (lower panel).
FIG. 5.
Validation of the potential target mRNAs by real-time PCR analysis. (A) Total RNAs were isolated from the translationally active polysomal fraction and inactive free fraction after 4 and 18 h of IFN-γ treatment of normal U937 cells. The total mRNAs from unfractionated cell lysates were also isolated at 0, 4, and 18 h of IFN-γ treatment to check the steady-state levels. Total RNAs were subjected to reverse transcription using a TaqMan Reverse Transcription Reagent Kit (Applied Biosystem, Foster City, CA). PCR amplification was carried out using Sybr Green PCR Master Mix (Applied Biosystem, Foster City, CA) and an ABI Thermo Cycler (ABI Prism 7000 SDS). In addition, the authenticity of each primer pair used in real-time PCR was confirmed by classical reverse transcription-PCR and sequencing of the amplified products (100 to 200 bp). The value shown is the relative amount of each mRNA after normalization with GAPDH from triplicates. (B) Status of polysomal and nonpolysomal abundances of two IFN-γ-induced and two noninduced mRNAs upon IFN-γ treatment as a negative control. Total RNAs were isolated from the translationally active polysomal fraction and inactive free fraction after 4 and 18 h of IFN-γ treatment of normal U937 cells. The total mRNAs from unfractionated cell lysates were also isolated at 0, 4, and 18 h of IFN-γ treatment to check the steady-state levels. Total RNAs were subjected to real-time PCR using a TaqMan Reverse Transcription Reagent Kit, Sybr Green PCR Master Mix, and an ABI Thermo Cycler (ABI Prism 7000 SDS). The value shown is the relative amount of each mRNA after normalization with GAPDH from triplicates. (C) The experiments described in panel A above were carried out using L13a-depleted cells expressing shRNA against L13a and control shRNA.
FIG. 6.
Abilities of the 3′ UTRs of target mRNAs to confer translational silencing to reporter mRNAs in response to IFN-γ treatment. (A) Normal U937 cells were electroporated using the construct harboring the CMV promoter-driven firefly Luc upstream of the different 3′ UTR elements under test and CMV-Renilla Luc construct as a control for transfection efficiencies. After the recovery period of the electroporated cells, they were treated with IFN-γ (500 U/ml) for 4 and 18 h. Chemiluminescence was measured to quantify the firefly and Renilla Luc activity. Renilla Luc expression was used to normalize the firefly Luc and then expressed as a percentage compared to untreated control cells. The data shown represent the means and standard errors from three independent experiments. (B) The experiment described above was conducted using L13a-depleted cells by stable expression of shRNA against L13a. (C) The experiment described above was conducted using cells expressing control shRNA. (D) Status of L13a depletion of the U937 cells used in the experiment. (E) Design of the reporter gene constructs harboring the elements from different 3′ UTRs at the end of the reporter Luc.
FIG. 7.
The asymmetric bulge or internal loop of CCL22 GAIT element is important for its activity. (A) Synthetic RNAs from the CCL22 wild-type (Wt) GAIT element (nt 433 to 462), CCL22 mutant (Mut) GAIT (A453 455C), and wild-type Cp GAIT (nt 78 to 106) were synthesized (Dharmacon) and 32P labeled by T4 polynucleotide kinase. The cell extracts from IFN-γ-treated U937 cells were incubated with 32P-labeled RNA. In competition experiments, the extracts were first incubated with 100- and 20-fold molar excesses of the unlabeled CCL22 wild-type GAIT element before the extract was added to the labeled probe. To test the requirement of L13a in the RNA protein complex formation, the extract was immunodepleted with L13a and control antibody before being added to the labeled probe. RNA protein complexes were resolved by electrophoresis on a nondenaturing 5% polyacrylamide gel and detected by autoradiography. (B) Reporter gene constructs were made harboring either the CCL22 wild-type GAIT element (nt 433 to 462) or mutant (MUT) GAIT element (A453 455C) at the end of the reporter Luc. These constructs were transfected in to U937 cells alongside the reporter construct harboring Cp GAIT (nt 78 to 106) as a positive control, followed by IFN-γ treatment for 4 and 18 h (for details, see the legend of Fig. 6A).
FIG. 8.
Detection of the steady-state levels of CCR3 and CXCL13 in IFN-γ-treated monocytes. U937 cells were treated with IFN-γ for different times. The cells were lysed and then subjected to by SDS-polyacrylamide gel electrophoresis analysis and immunoblotting using anti-CCR3 (Santa Cruz Biotechnology, Santa Cruz, CA) and anti-CXCL13 (R&D Systems, Minneapolis, MN).
FIG. 9.
GAIT complex-mediated translational silencing of the posttranscriptional operon controls inflammation in monocytes/macrophages. IFN-γ induces a cluster of mRNAs and synthesis of the encoded proteins involved in the inflammatory response; this process is proinflammatory. However, at 18 h posttreatment the formation of active GAIT complex causes the simultaneous translational silencing of this cluster by binding to the GAIT elements present in the 3′ UTRs of the target mRNAs. Using Cp mRNA, the first discovered member of this posttranscriptional operon, we have shown that the GAIT complex blocks the recruitment of the 40S ribosomal subunit (16). This process may resolve inflammation.
Similar articles
- Transcript-selective translational silencing by gamma interferon is directed by a novel structural element in the ceruloplasmin mRNA 3' untranslated region.
Sampath P, Mazumder B, Seshadri V, Fox PL. Sampath P, et al. Mol Cell Biol. 2003 Mar;23(5):1509-19. doi: 10.1128/MCB.23.5.1509-1519.2003. Mol Cell Biol. 2003. PMID: 12588972 Free PMC article. - Delayed translational silencing of ceruloplasmin transcript in gamma interferon-activated U937 monocytic cells: role of the 3' untranslated region.
Mazumder B, Fox PL. Mazumder B, et al. Mol Cell Biol. 1999 Oct;19(10):6898-905. doi: 10.1128/MCB.19.10.6898. Mol Cell Biol. 1999. PMID: 10490627 Free PMC article. - Conserved structures formed by heterogeneous RNA sequences drive silencing of an inflammation responsive post-transcriptional operon.
Basu A, Jain N, Tolbert BS, Komar AA, Mazumder B. Basu A, et al. Nucleic Acids Res. 2017 Dec 15;45(22):12987-13003. doi: 10.1093/nar/gkx979. Nucleic Acids Res. 2017. PMID: 29069516 Free PMC article. - Regulation of macrophage ceruloplasmin gene expression: one paradigm of 3'-UTR-mediated translational control.
Mazumder B, Sampath P, Fox PL. Mazumder B, et al. Mol Cells. 2005 Oct 31;20(2):167-72. Mol Cells. 2005. PMID: 16267389 Review. - The GAIT translational control system.
Arif A, Yao P, Terenzi F, Jia J, Ray PS, Fox PL. Arif A, et al. Wiley Interdiscip Rev RNA. 2018 Mar;9(2):e1441. doi: 10.1002/wrna.1441. Epub 2017 Nov 20. Wiley Interdiscip Rev RNA. 2018. PMID: 29152905 Free PMC article. Review.
Cited by
- Aminoacyl-tRNA Synthetase: A Non-Negligible Molecule in RNA Viral Infection.
Feng M, Zhang H. Feng M, et al. Viruses. 2022 Mar 15;14(3):613. doi: 10.3390/v14030613. Viruses. 2022. PMID: 35337020 Free PMC article. Review. - The role of eIF4F-driven mRNA translation in regulating the tumour microenvironment.
Bartish M, Abraham MJ, Gonçalves C, Larsson O, Rolny C, Del Rincón SV. Bartish M, et al. Nat Rev Cancer. 2023 Jun;23(6):408-425. doi: 10.1038/s41568-023-00567-5. Epub 2023 May 4. Nat Rev Cancer. 2023. PMID: 37142795 Review. - A viral pan-end RNA element and host complex define a SARS-CoV-2 regulon.
Khan D, Terenzi F, Liu G, Ghosh PK, Ye F, Nguyen K, China A, Ramachandiran I, Chakraborty S, Stefan J, Khan K, Vasu K, Dong F, Willard B, Karn J, Gack MU, Fox PL. Khan D, et al. Nat Commun. 2023 Jun 9;14(1):3385. doi: 10.1038/s41467-023-39091-3. Nat Commun. 2023. PMID: 37296097 Free PMC article. - Heterotrimeric GAIT complex drives transcript-selective translation inhibition in murine macrophages.
Arif A, Chatterjee P, Moodt RA, Fox PL. Arif A, et al. Mol Cell Biol. 2012 Dec;32(24):5046-55. doi: 10.1128/MCB.01168-12. Epub 2012 Oct 15. Mol Cell Biol. 2012. PMID: 23071094 Free PMC article. - Loss of function of ribosomal protein L13a blocks blastocyst formation and reveals a potential nuclear role in gene expression.
Kour R, Kim J, Roy A, Richardson B, Cameron MJ, Knott JG, Mazumder B. Kour R, et al. FASEB J. 2023 Dec;37(12):e23275. doi: 10.1096/fj.202301475R. FASEB J. 2023. PMID: 37902531 Free PMC article.
References
- Brown, V., P. Jin, S. Ceman, J. C. Darnell, W. T. O'Donnell, S. A. Tenenbaum, X. Jin, Y. Feng, K. D. Wilkinson, J. D. Keene, R. B. Darnell, and S. T. Warren. 2001. Microarray identification of FMRP-associated brain mRNAs and altered mRNA translational profiles in fragile X syndrome. Cell 107477-487. - PubMed
- Buxade, M., J. L. Parra, S. Rousseau, N. Shpiro, R. Marquez, N. Morrice, J. Bain, E. Espel, and C. G. Proud. 2005. The Mnks are novel components in the control of TNF alpha biosynthesis and phosphorylate and regulate hnRNP A1. Immunity 23177-189. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- HL79164/HL/NHLBI NIH HHS/United States
- AI068133/AI/NIAID NIH HHS/United States
- R01 AI059267/AI/NIAID NIH HHS/United States
- R01 HL079164/HL/NHLBI NIH HHS/United States
- R01 AI068133/AI/NIAID NIH HHS/United States
- AI059267/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Miscellaneous