Epigenetic regulation of centromeric chromatin: old dogs, new tricks? - PubMed (original) (raw)
Review
Epigenetic regulation of centromeric chromatin: old dogs, new tricks?
Robin C Allshire et al. Nat Rev Genet. 2008 Dec.
Abstract
The assembly of just a single kinetochore at the centromere of each sister chromatid is essential for accurate chromosome segregation during cell division. Surprisingly, despite their vital function, centromeres show considerable plasticity with respect to their chromosomal locations and activity. The establishment and maintenance of centromeric chromatin, and therefore the location of kinetochores, is epigenetically regulated. The histone H3 variant CENP-A is the key determinant of centromere identity and kinetochore assembly. Recent studies have identified many factors that affect CENP-A localization, but their precise roles in this process are unknown. We build on these advances and on new information about the timing of CENP-A assembly during the cell cycle to propose new models for how centromeric chromatin is established and propagated.
Figures
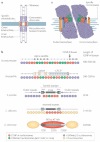
Figure 1. Centromere structure and organization
a | Centromeric chromatin underlies the kinetochore, which contains inner and outer plates that form microtubule-attachment sites. Pericentromeric heterochromatin flanks centromeric chromatin, and contains a high density of cohesin, which mediates sister-chromatid cohesion. b | Schematic depiction of centromeric DNA in humans and mice, Drosophila, Schizosaccharomyces pombe, Candida albicans and Saccharomyces cerevisiae. c | Model of the three-dimensional structure of mitotic centromeres. Data from mammalian and Drosophila cells suggest that centromeric chromatin is folded so that nucleosomes that are modified by dimethylation of lysine 4 of histone H3 (H3K4me2) form a discrete domain internal to CENP-A nucleosomes, which are presented on the chromosome surface, allowing kinetochore assembly and association with spindle microtubules. CDE,centromere DNA element; H2AZ, histone H2AZ; imr, innermost repeats.
Figure 2. CENP-A structure and nucleosome composition
a | The histone fold domain of histone H3 proteins is composed of four alpha helical domains (N and 1–3). Loop 1 separates 1 and 2. The CENP-A targeting domain (CATD) contains key differences from H3, and is sufficient for localization to centromeres when substituted into canonical H3 (the amino acids highlighted in orange are required in Drosophila). The CATD forms a rigid conformation in combination with H4, and this distinguishes CENP-A–H4 from H3–H4 heteromers. b | In non-centromeric regions, canonical histone H3 assembles into octameric nucleosomes composed of two H2A, H2B, H3, and H4 histone subunits. In centromeric chromatin, CENP-A can assemble into homotypic octamers, in which both H3 subunits are replaced by CENP-A, or into heterotypic octamers, which contain one canonical H3 and one CENP-A subunit. In Drosophila, CENP-A has been reported to form half-nucleosomes, that is, homotypic tetramers containing one subunit each of H2A, H2B, H4 and CENP-ACID (the Drosophila homologue of CENP-A). In Saccharomyces cerevisiae, Scm3 has been reported to replace the H2A–H2B dimers, forming an unusual hexamer containing two Scm3, CENP-A and H4 subunits. Additional forms of CENP-A nucleosomes can be envisaged by the replacement of one or both H2A subunits with H2A variants.
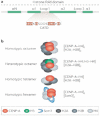
Figure 3. Cell-cycle regulation of CENP-A assembly
a | Pre-existing nucleosomes are distributed equally to sister-chromatids at the replication fork. The chromatin assembly factors CAF and ASF1 coordinate the deposition of new histone H3 and H4, and assemble new nucleosomes behind the replication fork. CENP-A density at centromeres drops by one half following replication, most probably owing to CENP-A-nucleosome segregation at the fork. The resulting nucleosome ‘gaps’ could be ‘filled’ with new H3 nucleosomes during S phase, or might be maintained as gaps until new CENP-A nucleosomes are assembled. CENP-A nucleosomes could also be split during replication, forming half-nucleosomes (not shown). b | Pulse–chase experiments in human cells show that new CENP-A is assembled at centromeres starting in telophase and continuing through G1. Thus, the amount of CENP-A at each centromere is highest at the end of G1, and is halved during S phase owing to the distribution of CENP-A to both sister chromatids. Cellular CENP-A protein levels are highest in G2, but new CENP-A assembly does not initiate until telophase. The amount of CENP-A in centromeres does not return to maximal levels until the end of G1, suggesting that kinetochore assembly occurs at centromeres containing half the maximal amount of CENP-A. MCM, minichromosome maintenance helicase; PCNA, proliferating cell nuclear antigen; DNA Pol, DNA polymerase.
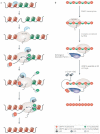
Figure 3. Cell-cycle regulation of CENP-A assembly
a | Nucleosome assembly pathways. Human and Saccharomyces cerevisiae proteins that are thought to function in these pathways are shown. Newly synthesized histone H3 and H4 are acetylated by a histone acetylase (HAT) and form tetramers in the cytoplasm. Chaperones, such as the human RbAp46/48 and NASP, facilitate H3–H4 import into the nucleus in combination with an importin protein, such as the S. cerevisiae Kap123. Replication-coupled assembly into chromatin is mediated in humans by the chromatin assembly factors ASF1 and CAF (which includes RbAp46/48), and acetyl group (Ac) modifications are removed by histone deacetylases. The protein complex HIRA mediates transcription-coupled assembly of H3–H4 into chromatin. Nucleoplasmin (in humans) and Nap1 (in S. cerevisiae) act as chaperones for H2A–H2B dimers and, in S. cerevisiae, Kap114 mediates their nuclear import. NASP and RbAp46/48 also associate with CENP-A, and might participate in its nuclear import. Specific post-translational modifications might also play a part in CENP-A nuclear import and deposition. The factors that mediate CENP-A assembly into chromatin are unknown. However, the facilitates chromatin transcription (FACT) remodelling complex associates with CENP-A, and RbAp46/48 can mediate CENP-A nucleosome assembly in vitro and also associates with the MIS18 complex, which is recruited to centromeres in telophase and through G1. Defects in histone gene transcription, translation, modification or import could affect the ability to assemble intact CENP-A chromatin and result in loss of CENP-A from centromeres (Box 1). b | Factors involved in CENP-A localization. Pericentromeric heterochromatin facilitates the establishment CENP-A on naked DNA templates in Schizosaccharomyces pombe, and the alpha satellite-binding protein CENP-B facilitates de novo centromere formation in human cells. Normally, propagation of centromere identity after DNA replication requires assembly of newly synthesized CENP-A at pre-existing locations. Factors that are physically associated with CENP-A include the nucleosome associated complex (NAC) and the MIS18 complex, RbAp46/48, NASP and FACT, among others. Some NAC components are reciprocally required for the centromere localization of CENP-A, demonstrating interdependency at the top of the propagation pathway. MIS18 components are known to function upstream of CENP-A, as their localization is not disrupted in CENP-A mutants. Whether these and/or other factors are specifically required for CENP-A assembly into chromatin is currently unknown. Other kinetochore components, such as MIS12, have been shown to localize independently of CENP-A. Components of the CENP-A distal (CAD) complex are not required for CENP-A localization, but CENP-A is required to recruit CAD and kinetochore components that assemble the inner and outer plates and microtubule attachment sites.
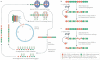
Figure 5. Possible roles for transcription of centromeric repeats
a | Transcription of centromeric chromatin might facilitate the replacement of histone H3 with CENP-A following replication (for example, in the subsequent G1 phase in human cells). The FACT (facilitates chromatin transcription) complex translocates nucleosomes from in front of RNA polymerase II (RNAPII) to behind it during transcription. FACT associates with CENP-A nucleosomes, suggesting that the act of transcription might be involved in the replacement of H3 with CENP-A, analogous to the transcription-coupled replacement of H3 with H3.3, which is mediated by HIRA. b | Non-coding RNAs that are homologous to centromeric DNA, that is, nascent transcripts or processed small RNAs (not shown), could have an active role in recruiting specific chromatin remodelling and/or assembly factors that direct CENP-A nucleosome deposition to centromeres. The deposition of H3 or the formation of nucleosomal gaps in centromeric chromatin during S phase could trigger transcription of centromeric DNA, and restoration of full levels of CENP-A nucleosomes could terminate assembly by blocking transcription.
Figure 6. Models for regulation of CENP-A propagation during the cell cycle
a | In human cells, new CENP-A is deposited at centromeres in G1 to restore maximum occupancy at centromeres before replication. In S phase, following replication, half of these CENP-A nucleosomes remain with each sister chromatid, and the spaces are either temporarily filled with H3-containing nucleosomes or remain as nucleosomal gaps until the end of mitosis (M). Successful kinetochore assembly, microtubule attachment, bi-orientation and segregation of chromosomes to opposite spindle poles during mitosis might somehow authorise CENP-A deposition in the following G1 phase. Restoration of CENP-A levels at centromeres might occur by recruitment of CENP-A assembly factors that deposit new CENP-A in gaps, or by exchange factors that mediate replacement of H3 nucleosomes with CENP-A nucleosomes. b | Specificity of CENP-A deposition mediated by the interspersed H3 blocks. Following replication of centromeric chromatin, a distinct combination of post-translational modifications in the interspersed H3 blocks (for example, dimethylation of lysine 4 of histone H3, H3K4me2) might specify the recruitment of factors that are required for the replacement of H3 with CENP-A exchange or the deposition of new CENP-A nucleosomes into nucleosomal gaps. c | Specificity of CENP-A deposition mediated by CENP-A. CENP-A octamers might split during replication so that half-nucleosomes composed of single H2A, H2B, CENP-A and H4 subunits are retained on each sister chromatid through G2 and metaphase. In telophase and through G1, half-nucleosomes might attract specific assembly factors that rebuild complete CENP-A octamers, thus restoring centromeric chromatin integrity. Alternatively, CENP-A octamers might be segregated intact at the replication fork, and might specify recruitment of factors that mediate assembly of CENP-A nucleosomes into adjacent gaps, or replacement of adjacent H3 nucleosomes deposited in S phase (not shown).
Similar articles
- Plasticity and epigenetic inheritance of centromere-specific histone H3 (CENP-A)-containing nucleosome positioning in the fission yeast.
Yao J, Liu X, Sakuno T, Li W, Xi Y, Aravamudhan P, Joglekar A, Li W, Watanabe Y, He X. Yao J, et al. J Biol Chem. 2013 Jun 28;288(26):19184-96. doi: 10.1074/jbc.M113.471276. Epub 2013 May 9. J Biol Chem. 2013. PMID: 23661703 Free PMC article. - How to build a centromere: from centromeric and pericentromeric chromatin to kinetochore assembly.
Vos LJ, Famulski JK, Chan GK. Vos LJ, et al. Biochem Cell Biol. 2006 Aug;84(4):619-39. doi: 10.1139/o06-078. Biochem Cell Biol. 2006. PMID: 16936833 Review. - The CENP-A nucleosome: a dynamic structure and role at the centromere.
Quénet D, Dalal Y. Quénet D, et al. Chromosome Res. 2012 Jul;20(5):465-79. doi: 10.1007/s10577-012-9301-4. Chromosome Res. 2012. PMID: 22825424 Free PMC article. Review. - Putting CENP-A in its place.
Stellfox ME, Bailey AO, Foltz DR. Stellfox ME, et al. Cell Mol Life Sci. 2013 Feb;70(3):387-406. doi: 10.1007/s00018-012-1048-8. Epub 2012 Jun 23. Cell Mol Life Sci. 2013. PMID: 22729156 Free PMC article. Review. - The centromere: chromatin foundation for the kinetochore machinery.
Fukagawa T, Earnshaw WC. Fukagawa T, et al. Dev Cell. 2014 Sep 8;30(5):496-508. doi: 10.1016/j.devcel.2014.08.016. Dev Cell. 2014. PMID: 25203206 Free PMC article. Review.
Cited by
- The Mub1/Ubr2 ubiquitin ligase complex regulates the conserved Dsn1 kinetochore protein.
Akiyoshi B, Nelson CR, Duggan N, Ceto S, Ranish JA, Biggins S. Akiyoshi B, et al. PLoS Genet. 2013;9(2):e1003216. doi: 10.1371/journal.pgen.1003216. Epub 2013 Feb 7. PLoS Genet. 2013. PMID: 23408894 Free PMC article. - CenH3 evolution in diploids and polyploids of three angiosperm genera.
Masonbrink RE, Gallagher JP, Jareczek JJ, Renny-Byfield S, Grover CE, Gong L, Wendel JF. Masonbrink RE, et al. BMC Plant Biol. 2014 Dec 30;14:383. doi: 10.1186/s12870-014-0383-3. BMC Plant Biol. 2014. PMID: 25547313 Free PMC article. - Chromatin and DNA replication.
MacAlpine DM, Almouzni G. MacAlpine DM, et al. Cold Spring Harb Perspect Biol. 2013 Aug 1;5(8):a010207. doi: 10.1101/cshperspect.a010207. Cold Spring Harb Perspect Biol. 2013. PMID: 23751185 Free PMC article. Review. - Quantitative single-molecule microscopy reveals that CENP-A(Cnp1) deposition occurs during G2 in fission yeast.
Lando D, Endesfelder U, Berger H, Subramanian L, Dunne PD, McColl J, Klenerman D, Carr AM, Sauer M, Allshire RC, Heilemann M, Laue ED. Lando D, et al. Open Biol. 2012 Jul;2(7):120078. doi: 10.1098/rsob.120078. Open Biol. 2012. PMID: 22870388 Free PMC article. - Structural transitions of centromeric chromatin regulate the cell cycle-dependent recruitment of CENP-N.
Fang J, Liu Y, Wei Y, Deng W, Yu Z, Huang L, Teng Y, Yao T, You Q, Ruan H, Chen P, Xu RM, Li G. Fang J, et al. Genes Dev. 2015 May 15;29(10):1058-73. doi: 10.1101/gad.259432.115. Epub 2015 May 5. Genes Dev. 2015. PMID: 25943375 Free PMC article.
References
- Hassold T, Hunt P. To err (meiotically) is human: the genesis of human aneuploidy. Nature Rev. Genet. 2001;2:280–291. - PubMed
- Monaco ZL, Moralli D. Progress in artificial chromosome technology. Biochem. Soc. Trans. 2006;34:324–327. - PubMed
- Westermann S, Drubin DG, Barnes G. Structures and functions of yeast kinetochore complexes. Annu. Rev. Biochem. 2007;76:563–591. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 065061/WT_/Wellcome Trust/United Kingdom
- R01 GM066272/GM/NIGMS NIH HHS/United States
- R01 GM066272-05/GM/NIGMS NIH HHS/United States
- 065061/Z/WT_/Wellcome Trust/United Kingdom
LinkOut - more resources
Full Text Sources