Autofix for backward-fit sidechains: using MolProbity and real-space refinement to put misfits in their place - PubMed (original) (raw)
Autofix for backward-fit sidechains: using MolProbity and real-space refinement to put misfits in their place
Jeffrey J Headd et al. J Struct Funct Genomics. 2009 Mar.
Abstract
Misfit sidechains in protein crystal structures are a stumbling block in using those structures to direct further scientific inference. Problems due to surface disorder and poor electron density are very difficult to address, but a large class of systematic errors are quite common even in well-ordered regions, resulting in sidechains fit backwards into local density in predictable ways. The MolProbity web site is effective at diagnosing such errors, and can perform reliable automated correction of a few special cases such as 180 degrees flips of Asn or Gln sidechain amides, using all-atom contacts and H-bond networks. However, most at-risk residues involve tetrahedral geometry, and their valid correction requires rigorous evaluation of sidechain movement and sometimes backbone shift. The current work extends the benefits of robust automated correction to more sidechain types. The Autofix method identifies candidate systematic, flipped-over errors in Leu, Thr, Val, and Arg using MolProbity quality statistics, proposes a corrected position using real-space refinement with rotamer selection in Coot, and accepts or rejects the correction based on improvement in MolProbity criteria and on chi angle change. Criteria are chosen conservatively, after examining many individual results, to ensure valid correction. To test this method, Autofix was run and analyzed for 945 representative PDB files and on the 50S ribosomal subunit of file 1YHQ. Over 40% of Leu, Val, and Thr outliers and 15% of Arg outliers were successfully corrected, resulting in a total of 3,679 corrected sidechains, or 4 per structure on average. Summary Sentences: A common class of misfit sidechains in protein crystal structures is due to systematic errors that place the sidechain backwards into the local electron density. A fully automated method called "Autofix" identifies such errors for Leu, Val, Thr, and Arg and corrects over one third of them, using MolProbity validation criteria and Coot real-space refinement of rotamers.
Figures
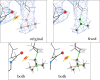
Fig. 1
Example Autofix correction of a Leu decoy rotamer from the 945-file dataset: Leu D 427 from 1A0E (Thermotoga neapolitana xylose isomerase) at 2.7 Å resolution. a (original) Leu D 427 in its deposited conformation, which is a rotamer outlier with an eclipsed χ angle and a clash with Leu D 430. b (both) Overlay, in stereo, of proposed corrected Leu rotamer (green) over the deposited conformation (pink). c (fixed) Corrected Leu D 427, in a favored mt rotamer. The clash with Leu D 430 has been alleviated and the bond angle idealized, with a somewhat better fit to the density. Images in Figs. 1, 2 and 4 were generated using KING [3]
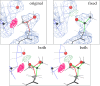
Fig. 2
Example Autofix correction from the 50S ribosome: a Thr rotamer outlier, from protein L18e in the 1YHQ archaeal large ribosomal subunit (2.4 Å) [15], before and after correction. a (original) Thr O 3 in its deposited orientation, with fairly good fit to the density, but a serious clash with RNA backbone (Thr methyl to G 0 656 H5′), no H-bond, and a rotamer outlier. b (both) Overlay, in stereo, of proposed corrected Thr rotamer (green) over the original position (pink). c (fixed) Corrected Thr O 3, with equivalent fit to the density, no steric clashes, an excellent p rotamer, and now a strong H-bond from Thr OG1 to the 2′OH of G 0 655. C atoms are gray or black balls; O atoms are larger red balls. Steric clashes are shown as clusters of hot pink spikes, H-bonds as lenses of pale green dots
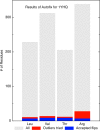
Fig. 3
Summary of Autofix results on 1YHQ 50S ribosomal subunit. Bar chart summary of correction results on Leu, Thr, Val, and Arg residues in 1YHQ. Gray bars represent the total number of each residue type in the file. Red represents the number of candidate outliers (<1% rotamer score). Blue represents the number of successfully corrected residues of each type: 7 Leu, 8 Val, 8 Thr, and 7 Arg, which are 63, 57, 67, and 25% of the outliers, respectively
Fig. 4
Before and after χ1–χ2 plots of the 2,037 accepted Leu corrections, for those identified as rotamer outliers (<1%) in our 945-file dataset and successfully corrected by Autofix. Contours are taken from the Top500 Leu set [1], with decoys removed; black lines are the 1% contours and gray lines are the 10% contours of rotamer score. a Before: χ1–χ2 plot for the original conformation of each corrected Leu outlier (thus outside the 1% contours). b After: χ1–χ2 plot of the final χ values for each successfully corrected Leu outlier (now inside the contours). Data points are color-coded by which rotamer they ended up in after correction: mt green, tp blue, tt red, mp brown, pp purple, tm yellow, mm hot pink, and pt orange. Note that for most rotamers, the corrected examples came from a well-defined decoy cluster approximately 180° distant
Fig. 5
Summary of real-space correlation coefficients (RSCC) for corrected outlier residues before (gray) and after (black) Autofix, showing improvement for all 4 amino acid types. Median RSCC values are indicated by a vertical line. The box around the median spans the 25th to the 75th percentile. Whiskers end at the 1st or 99th percentile
Similar articles
- MolProbity: More and better reference data for improved all-atom structure validation.
Williams CJ, Headd JJ, Moriarty NW, Prisant MG, Videau LL, Deis LN, Verma V, Keedy DA, Hintze BJ, Chen VB, Jain S, Lewis SM, Arendall WB 3rd, Snoeyink J, Adams PD, Lovell SC, Richardson JS, Richardson DC. Williams CJ, et al. Protein Sci. 2018 Jan;27(1):293-315. doi: 10.1002/pro.3330. Epub 2017 Nov 27. Protein Sci. 2018. PMID: 29067766 Free PMC article. - NQ-Flipper: recognition and correction of erroneous asparagine and glutamine side-chain rotamers in protein structures.
Weichenberger CX, Sippl MJ. Weichenberger CX, et al. Nucleic Acids Res. 2007 Jul;35(Web Server issue):W403-6. doi: 10.1093/nar/gkm263. Epub 2007 May 3. Nucleic Acids Res. 2007. PMID: 17478502 Free PMC article. - New tools in MolProbity validation: CaBLAM for CryoEM backbone, UnDowser to rethink "waters," and NGL Viewer to recapture online 3D graphics.
Prisant MG, Williams CJ, Chen VB, Richardson JS, Richardson DC. Prisant MG, et al. Protein Sci. 2020 Jan;29(1):315-329. doi: 10.1002/pro.3786. Epub 2019 Dec 10. Protein Sci. 2020. PMID: 31724275 Free PMC article. - You are lost without a map: Navigating the sea of protein structures.
Lamb AL, Kappock TJ, Silvaggi NR. Lamb AL, et al. Biochim Biophys Acta. 2015 Apr;1854(4):258-68. doi: 10.1016/j.bbapap.2014.12.021. Epub 2014 Dec 29. Biochim Biophys Acta. 2015. PMID: 25554228 Free PMC article. Review. - New Biological Insights from Better Structure Models.
Touw WG, Joosten RP, Vriend G. Touw WG, et al. J Mol Biol. 2016 Mar 27;428(6):1375-1393. doi: 10.1016/j.jmb.2016.02.002. Epub 2016 Feb 8. J Mol Biol. 2016. PMID: 26869101 Review.
Cited by
- Fitmunk: improving protein structures by accurate, automatic modeling of side-chain conformations.
Porebski PJ, Cymborowski M, Pasenkiewicz-Gierula M, Minor W. Porebski PJ, et al. Acta Crystallogr D Struct Biol. 2016 Feb;72(Pt 2):266-80. doi: 10.1107/S2059798315024730. Epub 2016 Jan 28. Acta Crystallogr D Struct Biol. 2016. PMID: 26894674 Free PMC article. - Local Protein Structure Refinement via Molecular Dynamics Simulations with locPREFMD.
Feig M. Feig M. J Chem Inf Model. 2016 Jul 25;56(7):1304-12. doi: 10.1021/acs.jcim.6b00222. Epub 2016 Jul 13. J Chem Inf Model. 2016. PMID: 27380201 Free PMC article. - Refining the macromolecular model - achieving the best agreement with the data from X-ray diffraction experiment.
Shabalin IG, Porebski PJ, Minor W. Shabalin IG, et al. Crystallogr Rev. 2018;24(4):236-262. doi: 10.1080/0889311X.2018.1521805. Epub 2018 Sep 21. Crystallogr Rev. 2018. PMID: 30416256 Free PMC article. - The Phenix software for automated determination of macromolecular structures.
Adams PD, Afonine PV, Bunkóczi G, Chen VB, Echols N, Headd JJ, Hung LW, Jain S, Kapral GJ, Grosse Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner RD, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. Adams PD, et al. Methods. 2011 Sep;55(1):94-106. doi: 10.1016/j.ymeth.2011.07.005. Epub 2011 Jul 29. Methods. 2011. PMID: 21821126 Free PMC article. - Intrinsic α-helical and β-sheet conformational preferences: a computational case study of alanine.
Caballero D, Määttä J, Zhou AQ, Sammalkorpi M, O'Hern CS, Regan L. Caballero D, et al. Protein Sci. 2014 Jul;23(7):970-80. doi: 10.1002/pro.2481. Epub 2014 May 9. Protein Sci. 2014. PMID: 24753338 Free PMC article.
References
- {'text': '', 'ref_index': 1, 'ids': [{'type': 'DOI', 'value': '10.1002/1097-0134(20000815)40:3<389::AID-PROT50>3.0.CO;2-2', 'is_inner': False, 'url': 'https://doi.org/10.1002/1097-0134(20000815)40:3<389::aid-prot50>3.0.co;2-2'}, {'type': 'PubMed', 'value': '10861930', 'is_inner': True, 'url': 'https://pubmed.ncbi.nlm.nih.gov/10861930/'}\]}
- Lovell SC, Word JM, Richardson JS, Richardson DC (2000) Proteins 40:389–408. doi:10.1002/1097-0134(20000815)40:3<389::AID-PROT50>3.0.CO;2-2 - PubMed
- {'text': '', 'ref_index': 1, 'ids': [{'type': 'DOI', 'value': '10.1006/jmbi.1998.2401', 'is_inner': False, 'url': 'https://doi.org/10.1006/jmbi.1998.2401'}, {'type': 'PubMed', 'value': '9917408', 'is_inner': True, 'url': 'https://pubmed.ncbi.nlm.nih.gov/9917408/'}\]}
- Word JM, Lovell SC, Richardson JS, Richardson DC (1999) J Mol Biol 285:1735–1747. doi:10.1006/jmbi.1998.2401 - PubMed
- {'text': '', 'ref_index': 1, 'ids': [{'type': 'DOI', 'value': '10.1093/nar/gkm216', 'is_inner': False, 'url': 'https://doi.org/10.1093/nar/gkm216'}, {'type': 'PMC', 'value': 'PMC1933162', 'is_inner': False, 'url': 'https://pmc.ncbi.nlm.nih.gov/articles/PMC1933162/'}, {'type': 'PubMed', 'value': '17452350', 'is_inner': True, 'url': 'https://pubmed.ncbi.nlm.nih.gov/17452350/'}\]}
- Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, Wang W, Murray LW, Arendall WBIII, Snoeyink J, Richardson JS, Richardson DC (2007) Nucleic Acids Res 35:W375–W383. doi:10.1093/nar/gkm216 - PMC - PubMed
- {'text': '', 'ref_index': 1, 'ids': [{'type': 'DOI', 'value': '10.1007/s10969-005-3138-4', 'is_inner': False, 'url': 'https://doi.org/10.1007/s10969-005-3138-4'}, {'type': 'PubMed', 'value': '15965733', 'is_inner': True, 'url': 'https://pubmed.ncbi.nlm.nih.gov/15965733/'}\]}
- Arendall BWIII, Tempel W, Richardson JS, Zhou W, Wang S, Davis IW, Liu Z-J, Rose JP, Carson WM, Luo M, Richardson DC, Wang B-C (2005) J Struct Funct Genomics 6:1–11. doi:10.1007/s10969-005-3138-4 - PubMed
- {'text': '', 'ref_index': 1, 'ids': [{'type': 'DOI', 'value': '10.1093/nar/28.1.235', 'is_inner': False, 'url': 'https://doi.org/10.1093/nar/28.1.235'}, {'type': 'PMC', 'value': 'PMC102472', 'is_inner': False, 'url': 'https://pmc.ncbi.nlm.nih.gov/articles/PMC102472/'}, {'type': 'PubMed', 'value': '10592235', 'is_inner': True, 'url': 'https://pubmed.ncbi.nlm.nih.gov/10592235/'}\]}
- Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE (2000) Nucleic Acids Res 28:235–242. doi:10.1093/nar/28.1.235 - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM073930/GM/NIGMS NIH HHS/United States
- GM073919/GM/NIGMS NIH HHS/United States
- GM074127/GM/NIGMS NIH HHS/United States
- R01 GM074127/GM/NIGMS NIH HHS/United States
- R01 GM073919-04/GM/NIGMS NIH HHS/United States
- P01 GM063210/GM/NIGMS NIH HHS/United States
- R01 GM073930-04S1/GM/NIGMS NIH HHS/United States
- R01 GM073919/GM/NIGMS NIH HHS/United States
- GM063210/GM/NIGMS NIH HHS/United States
- GM073930/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources