The influence of sex, handedness, and washing on the diversity of hand surface bacteria - PubMed (original) (raw)
The influence of sex, handedness, and washing on the diversity of hand surface bacteria
Noah Fierer et al. Proc Natl Acad Sci U S A. 2008.
Abstract
Bacteria thrive on and within the human body. One of the largest human-associated microbial habitats is the skin surface, which harbors large numbers of bacteria that can have important effects on health. We examined the palmar surfaces of the dominant and nondominant hands of 51 healthy young adult volunteers to characterize bacterial diversity on hands and to assess its variability within and between individuals. We used a novel pyrosequencing-based method that allowed us to survey hand surface bacterial communities at an unprecedented level of detail. The diversity of skin-associated bacterial communities was surprisingly high; a typical hand surface harbored >150 unique species-level bacterial phylotypes, and we identified a total of 4,742 unique phylotypes across all of the hands examined. Although there was a core set of bacterial taxa commonly found on the palm surface, we observed pronounced intra- and interpersonal variation in bacterial community composition: hands from the same individual shared only 17% of their phylotypes, with different individuals sharing only 13%. Women had significantly higher diversity than men, and community composition was significantly affected by handedness, time since last hand washing, and an individual's sex. The variation within and between individuals in microbial ecology illustrated by this study emphasizes the challenges inherent in defining what constitutes a "healthy" bacterial community; addressing these challenges will be critical for the International Human Microbiome Project.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
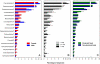
Fig. 1.
Relative abundances of the most abundant bacterial groups on the hand surfaces, with the hand samples divided into categories of sex (A), time since last hand washing (B), and the dominant versus the nondominant hand (C). Error bars are 1 standard error of the mean. For the number of sequences and number of samples included in each category and the full taxonomic description of the hand surface bacterial communities see
Table S1
. Superscripts on the taxon name indicate the phylum or subphylum: 1, Actinobacteria; 2, Firmicutes; 3, Betaproteobacteria; 4, Gammaproteobacteria; 5, Alphaproteobacteria; 6, Bacteroidetes.
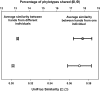
Fig. 2.
Average pairwise bacterial community similarity between left and right hands from the same individual (circles) and between hands from different individuals (squares) as measured by using the unweighted UniFrac similarity index (bottom axis, open symbols) or the percentage of phylotypes that are shared between pairs (top axis, filled symbols). Average pairwise values and 95% confidence intervals are shown. For these analyses, 2,500 sequences were randomly selected per sample, and only those samples represented by >2,500 sequences were included (n = 51 and 5,100 pairwise comparisons for intraindividual comparison and interindividual comparisons, respectively).
Fig. 3.
Differentiation in hand-surface communities between sexes (A), dominant versus the nondominant hands (B), time since last hand washing (C), and time since last hand washing for each sex (D) determined by using the unweighted UniFrac algorithm. The length of the branches corresponds to the degree of differentiation between bacterial communities in each category. All of the branch nodes shown here were found to be significant (P < 0.001), indicating that each of these categories harbored distinct bacterial communities.
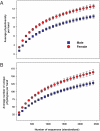
Fig. 4.
Rarefaction curves showing differences in bacterial diversity on palm surfaces from men and women. (A) Phylogenetic diversity estimated by measuring the average total branch length per sample after a specified number of individual sequences have been observed (36). (B) Diversity estimated by determining the average number of unique phylotypes per hand. For these analyses, we randomly selected 2,400 sequences per hand sample, and thus the average number of phylotypes per hand is lower than for the full dataset (Table 1). Bars indicate 95% confidence intervals.
Similar articles
- Hand bacterial communities vary across two different human populations.
Hospodsky D, Pickering AJ, Julian TR, Miller D, Gorthala S, Boehm AB, Peccia J. Hospodsky D, et al. Microbiology (Reading). 2014 Jun;160(Pt 6):1144-1152. doi: 10.1099/mic.0.075390-0. Epub 2014 May 11. Microbiology (Reading). 2014. PMID: 24817404 - Temporal variability is a personalized feature of the human microbiome.
Flores GE, Caporaso JG, Henley JB, Rideout JR, Domogala D, Chase J, Leff JW, Vázquez-Baeza Y, Gonzalez A, Knight R, Dunn RR, Fierer N. Flores GE, et al. Genome Biol. 2014 Dec 3;15(12):531. doi: 10.1186/s13059-014-0531-y. Genome Biol. 2014. PMID: 25517225 Free PMC article. - The method used to dry washed hands affects the number and type of transient and residential bacteria remaining on the skin.
Mutters R, Warnes SL. Mutters R, et al. J Hosp Infect. 2019 Apr;101(4):408-413. doi: 10.1016/j.jhin.2018.12.005. Epub 2018 Dec 8. J Hosp Infect. 2019. PMID: 30537524 - Hand-to-surface bacterial transfer and healthcare-associated infections prevention: a pilot study on skin microbiome in a molecular biology laboratory.
Delicati A, Marcante B, Catelan D, Biggeri A, Caenazzo L, Tozzo P. Delicati A, et al. Front Med (Lausanne). 2025 Mar 21;12:1546298. doi: 10.3389/fmed.2025.1546298. eCollection 2025. Front Med (Lausanne). 2025. PMID: 40190580 Free PMC article. - Review of human hand microbiome research.
Edmonds-Wilson SL, Nurinova NI, Zapka CA, Fierer N, Wilson M. Edmonds-Wilson SL, et al. J Dermatol Sci. 2015 Oct;80(1):3-12. doi: 10.1016/j.jdermsci.2015.07.006. Epub 2015 Jul 23. J Dermatol Sci. 2015. PMID: 26278471 Review.
Cited by
- Risk factors for cardiac implantable electronic device infections: a nationwide Danish study.
Olsen T, Jørgensen OD, Nielsen JC, Thøgersen AM, Philbert BT, Frausing MHJP, Sandgaard NCF, Johansen JB. Olsen T, et al. Eur Heart J. 2022 Dec 14;43(47):4946-4956. doi: 10.1093/eurheartj/ehac576. Eur Heart J. 2022. PMID: 36263789 Free PMC article. - Increased efficiency in identifying mixed pollen samples by meta-barcoding with a dual-indexing approach.
Sickel W, Ankenbrand MJ, Grimmer G, Holzschuh A, Härtel S, Lanzen J, Steffan-Dewenter I, Keller A. Sickel W, et al. BMC Ecol. 2015 Jul 22;15:20. doi: 10.1186/s12898-015-0051-y. BMC Ecol. 2015. PMID: 26194794 Free PMC article. - Biogeography of the ecosystems of the healthy human body.
Zhou Y, Gao H, Mihindukulasuriya KA, La Rosa PS, Wylie KM, Vishnivetskaya T, Podar M, Warner B, Tarr PI, Nelson DE, Fortenberry JD, Holland MJ, Burr SE, Shannon WD, Sodergren E, Weinstock GM. Zhou Y, et al. Genome Biol. 2013 Jan 14;14(1):R1. doi: 10.1186/gb-2013-14-1-r1. Genome Biol. 2013. PMID: 23316946 Free PMC article. - Biphasic assembly of the murine intestinal microbiota during early development.
Pantoja-Feliciano IG, Clemente JC, Costello EK, Perez ME, Blaser MJ, Knight R, Dominguez-Bello MG. Pantoja-Feliciano IG, et al. ISME J. 2013 Jun;7(6):1112-5. doi: 10.1038/ismej.2013.15. Epub 2013 Mar 28. ISME J. 2013. PMID: 23535917 Free PMC article. - Groundtruthing next-gen sequencing for microbial ecology-biases and errors in community structure estimates from PCR amplicon pyrosequencing.
Lee CK, Herbold CW, Polson SW, Wommack KE, Williamson SJ, McDonald IR, Cary SC. Lee CK, et al. PLoS One. 2012;7(9):e44224. doi: 10.1371/journal.pone.0044224. Epub 2012 Sep 6. PLoS One. 2012. PMID: 22970184 Free PMC article.
References
- Fredricks DN. Microbial ecology of human skin in health and disease. J Invest Dermatol Symp Proc. 2001;6:167–169. - PubMed
- Roth RR, James WD. Microbial ecology of the skin. Annu Rev Microbiol. 1988;42:441–464. - PubMed
Publication types
MeSH terms
Grants and funding
- P01 DK078669/DK/NIDDK NIH HHS/United States
- T32 GM065103/GM/NIGMS NIH HHS/United States
- P01DK078669/DK/NIDDK NIH HHS/United States
- T32GM065103/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases