microRNA-138 modulates cardiac patterning during embryonic development - PubMed (original) (raw)
microRNA-138 modulates cardiac patterning during embryonic development
Sarah U Morton et al. Proc Natl Acad Sci U S A. 2008.
Abstract
Organ patterning during embryonic development requires precise temporal and spatial regulation of protein activity. microRNAs (miRNAs), small noncoding RNAs that typically inhibit protein expression, are broadly important for proper development, but their individual functions during organogenesis are largely unknown. We report that miR-138 is expressed in specific domains in the zebrafish heart and is required to establish appropriate chamber-specific gene expression patterns. Disruption of miR-138 function led to ventricular expansion of gene expression normally restricted to the atrio-ventricular valve region and, ultimately, to disrupted ventricular cardiomyocyte morphology and cardiac function. Temporal-specific knockdown of miR-138 by antagomiRs showed miR-138 function was required during a discrete developmental window, 24-34 h post-fertilization (hpf). miR-138 functioned partially by repressing the retinoic acid synthesis enzyme, aldehyde dehydrogenase-1a2, in the ventricle. This activity was complemented by miR-138-mediated ventricular repression of the gene encoding versican (cspg2), which was positively regulated by retinoic-acid signaling. Our findings demonstrate that miR-138 helps establish discrete domains of gene expression during cardiac morphogenesis by targeting multiple members of a common pathway, and also establish the use of antagomiRs in fish for temporal knockdown of miRNA function.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
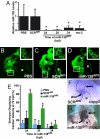
Fig. 1.
miR-138 is required for cardiac development. (A) Oblique view of 72 hpf fish after in situ hybridization showing expression of miR-138 in the ventricular chamber marked by blue staining. (B) qRT-PCR analysis of mature miR-138 expression at 48 hpf in flow-sorted cardiomyocytes (cmlc2:GFP), endothelial cells (flk1:GFP), or whole embryos demonstrated enrichment of miR-138 in cardiomyocytes. Mature miR-138 was not detected in endothelial cells. (C and D) Fish embryos injected with miR-138 scrambled control morpholino (scr) (C) or miR-138 morpholino (mo-1) (D) showing pericardial edema (asterisk); arrowhead indicates heart. (E–I) Representative confocal images of transgenic Tg(myl7:HRAS-mEGFP)s843 uninjected embryos (E) or embryos injected with 17-nt miR-138scr (F), 17-nt miR-138mo-1 (G), or a second 31-nt miR-138mo-2 oligo (H). Insets show single _z_-stack images of outer curvature ventricular myocytes, which normally elongate with maturation (E and F) but remained rounded in the miR-138mo embryos (G and H). (e, eye; y, yolk sac; a, atrium; v, ventricle; avc, atrioventricular canal.)
Fig. 2.
miR-138 knockdown leads to expansion of AVC-specific gene expression into the ventricle. (A–H) Ventral views of 48-hpf embryos after mRNA in situ hybridization focusing on head (h) and heart (dotted lines) regions. Atrial (A and B) and ventricular (C and D) markers were similar in wild-type (wt) (A and C) and miR-138mo-1 embryos (B and D). Expression of AVC-specific markers cspg2 (E and F) and notch1b (G and H) expanded into the ventricles (v) in miR-138mo-1 embryos (F and H) but not in wild-type embryos (E and G). (a, atrium; amhc, atrial myosin heavy chain; vmhc, ventricular myosin heavy chain.)
Fig. 3.
Temporal regulation of miRNA function by antagomiRs in zebrafish. (A) miR-138 RNA levels detected by qRT-PCR in 72-hpf fish embryos treated with antagomiRs from 24, 30, or 34 h postfertilization (hpf) or injected with the 31-nt morpholino (mo-2) compared with PBS- or scrambled antagomiR-treated controls. (B–D) Confocal images of hearts of 72-hpf Tg(myl7:HRAS-mEGFP)s843 transgenic embryos treated with PBS (B), scrambled antagomiR (SCRam) (C), or miR-138 antagomiR (miR-138am) (D) from 24–72 hpf. GFP revealed rounded myocyte morphology (Inset) in miR-138am embryos compared to the normally elongated myocytes seen in controls. (E) Pericardial edema indicating cardiac dysfunction and rounded ventricular myocytes reflecting delayed maturation were observed in varying percentages of 72-hpf embryos treated with PBS, SCRam, or miR-138am from the hpf indicated. (F and G) Ventral view of mRNA in situ hybridization to detect cspg2 expression in hearts of embryos treated with SCRam (F) or miR-138am (G) showed expansion of cspg2 into the ventricle of miR-138am embryos; heart is indicated with dotted lines. Results shown are the average of four experiments in (A) and (E). (v, ventricle; a, atrium; atrioventricular canal (avc); *, P < 0.05.)
Fig. 4.
miR-138 directly targets aldh1a2, restricting its expression to the AVC. (A) miR-138 binding site in zebrafish aldh1a2 3′ UTR with complementary nucleotides indicated in red. Luciferase activity in Cos cells on introduction of wild-type or mutated (mut) aldh1a2 3′ UTR sequences downstream of a CMV-driven luciferase reporter with or without miR-138 is shown. (B) aldh1a2 mRNA levels measured by qRT-PCR in embryos injected with pri-miR-138 or treated at 24 hpf with miR-138 antagomiR (miR-138am) or scrambled antagomiR (SCRam). Results shown represent at least 4 experiments in (A) and (B). Error bars indicate 95% confidence intervals. (C and D) Raldh2 immunohistochemistry on sections of 72-hpf embryos treated with PBS or miR-138am from 24–72 hpf. Ventricular (v) cardiomyocyte expression (arrowhead) was readily detectable in miR-138am hearts but not in controls, whereas intensity of staining in the AVC was similar. (E–G) Ventral view of cspg2 expression in the heart by mRNA in situ hybridization in embryos treated with scrambled morpholino (miR-138scr) (E), miR-138mo-1 (F), or retinoic acid (RA) (G); heart is indicated with dotted lines. (H–K) Confocal images of myl7-GFP embryos (48 hpf) treated with vehicle (DMSO) (H), premiR-138 RNA (I), or DEAB (J), and DEAB-treated embryos also injected with miR-138mo-1 (K) showing rescue of the linear heart tube phenotype with knockdown of miR-138. (L–N) Expansion of cspg2 mRNA expression from the avc (L) into the ventricle (v) of miR-138mo-1 embryos (M) was not rescued by DEAB (N). (a, atrium; *, P < 0.05.)
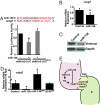
Fig. 5.
miR-138 directly targets cspg2. (A) Sequence of miR-138 and its binding site in mouse cspg2 3′ UTR with complementary nucleotides indicated in red. Luciferase activity in Cos cells on introduction of wild type or mutated (mut) cspg2 3′ UTR sequences downstream of a CMV-driven luciferase (LUC) reporter with or without miR-138 is shown. (B and C) Analysis of cspg2 mRNA level assessed by qRT-PCR (B) and corresponding versican protein by western blot (C) in mouse NIH 3T3 fibroblasts transfected with miR-138. (D) cspg2 mRNA levels detected by qRT-PCR in zebrafish embryos injected with miR-138 or treated at 24 hpf with miR-138 antagomiR (am) or scrambled (SCR) antagomiR. (E) Proposed model for the function of miR-138 in regulating chamber-specific gene expression and atrioventricular canal (avc) patterning. miR-138 directly represses retinoic acid (RA) synthesis in the ventricle (v) via aldh1a2, which would otherwise induce expression of the avc-specific gene, cspg2 (versican). Repression of cspg2 by miR-138 in the ventricle is also accomplished by directly targeting cspg2. Results shown in (A), (B), and (D) represent at least 4 experiments with error bars indicating 95% confidence intervals. (a, atrium; *, P < 0.05.)
Similar articles
- Pdlim7 (LMP4) regulation of Tbx5 specifies zebrafish heart atrio-ventricular boundary and valve formation.
Camarata T, Krcmery J, Snyder D, Park S, Topczewski J, Simon HG. Camarata T, et al. Dev Biol. 2010 Jan 15;337(2):233-45. doi: 10.1016/j.ydbio.2009.10.039. Epub 2009 Nov 3. Dev Biol. 2010. PMID: 19895804 Free PMC article. - miR-142-3p is essential for hematopoiesis and affects cardiac cell fate in zebrafish.
Nishiyama T, Kaneda R, Ono T, Tohyama S, Hashimoto H, Endo J, Tsuruta H, Yuasa S, Ieda M, Makino S, Fukuda K. Nishiyama T, et al. Biochem Biophys Res Commun. 2012 Sep 7;425(4):755-61. doi: 10.1016/j.bbrc.2012.07.148. Epub 2012 Aug 2. Biochem Biophys Res Commun. 2012. PMID: 22884798 - miR-196 regulates axial patterning and pectoral appendage initiation.
He X, Yan YL, Eberhart JK, Herpin A, Wagner TU, Schartl M, Postlethwait JH. He X, et al. Dev Biol. 2011 Sep 15;357(2):463-77. doi: 10.1016/j.ydbio.2011.07.014. Epub 2011 Jul 20. Dev Biol. 2011. PMID: 21787766 Free PMC article. - ZebRA: An overview of retinoic acid signaling during zebrafish development.
Samarut E, Fraher D, Laudet V, Gibert Y. Samarut E, et al. Biochim Biophys Acta. 2015 Feb;1849(2):73-83. doi: 10.1016/j.bbagrm.2014.05.030. Epub 2014 Jun 10. Biochim Biophys Acta. 2015. PMID: 24928143 Review. - MicroRNA regulatory networks in cardiovascular development.
Liu N, Olson EN. Liu N, et al. Dev Cell. 2010 Apr 20;18(4):510-25. doi: 10.1016/j.devcel.2010.03.010. Dev Cell. 2010. PMID: 20412767 Free PMC article. Review.
Cited by
- MicroRNAs and Cardiac Regeneration.
Hodgkinson CP, Kang MH, Dal-Pra S, Mirotsou M, Dzau VJ. Hodgkinson CP, et al. Circ Res. 2015 May 8;116(10):1700-11. doi: 10.1161/CIRCRESAHA.116.304377. Circ Res. 2015. PMID: 25953925 Free PMC article. Review. - Context-dependent functions of specific microRNAs in neuronal development.
Gao FB. Gao FB. Neural Dev. 2010 Oct 1;5:25. doi: 10.1186/1749-8104-5-25. Neural Dev. 2010. PMID: 20920300 Free PMC article. Review. - MicroRNAs in cardiac apoptosis.
Li P. Li P. J Cardiovasc Transl Res. 2010 Jun;3(3):219-24. doi: 10.1007/s12265-010-9175-9. Epub 2010 Mar 19. J Cardiovasc Transl Res. 2010. PMID: 20560043 Review. - microRNAs and Cardiac Cell Fate.
Piubelli C, Meraviglia V, Pompilio G, D'Alessandra Y, Colombo GI, Rossini A. Piubelli C, et al. Cells. 2014 Aug 5;3(3):802-23. doi: 10.3390/cells3030802. Cells. 2014. PMID: 25100020 Free PMC article. Review. - Illustrating the interplay between the extracellular matrix and microRNAs.
Piccinini AM, Midwood KS. Piccinini AM, et al. Int J Exp Pathol. 2014 Jun;95(3):158-80. doi: 10.1111/iep.12079. Epub 2014 Apr 25. Int J Exp Pathol. 2014. PMID: 24761792 Free PMC article. Review.
References
- Srivastava D. Making or breaking the heart: From lineage determination to morphogenesis. Cell. 2006;126:1037–1048. - PubMed
- Stainier DY. Zebrafish genetics and vertebrate heart formation. Nat Rev Genet. 2001;2:39–48. - PubMed
- Beis D, et al. Genetic and cellular analyses of zebrafish atrioventricular cushion and valve development. Development. 2005;132:4193–4204. - PubMed
- Yelon D, Horne SA, Stainier DY. Restricted expression of cardiac myosin genes reveals regulated aspects of heart tube assembly in zebrafish. Dev Biol. 1999;214:23–37. - PubMed
- Walsh EC, Stainier DY. UDP-glucose dehydrogenase required for cardiac valve formation in zebrafish. Science. 2001;293:1670–1673. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases