Regulation of hierarchical clustering and activation of innate immune cells by dendritic cells - PubMed (original) (raw)
Regulation of hierarchical clustering and activation of innate immune cells by dendritic cells
Suk-Jo Kang et al. Immunity. 2008.
Abstract
An early granulomatous response, characterized by collections of white blood cells at foci surrounding pathogens, occurs after infection by many intracellular organisms, including Listeria, but how these clusters become organized and for what purpose remain poorly understood. Here, we showed that dendritic cell (DC) activation by Listeria nucleated rapid clustering of innate cells, including granulocytes, natural killer (NK) cells, and monocytes, to sites of bacteria propagation where interleukin-12 was expressed in the spleen. Clustered NK cells expressed interferon-gamma (IFN-gamma), which was necessary for the activation and maturation of colocalized monocytes to tumor necrosis factor- and inducible nitric oxide synthase-producing DCs (TipDCs). NK cell clustering was necessary for IFN-gamma production and required pertussis-toxin-sensitive recruitment, in part mediated by the chemokine receptor CCR5, and MyD88 adaptor-mediated signaling. Thus, spatial organization of the immune response by DCs between 6 and 24 hr ensures functional activation of innate cells, which restricts pathogens before adaptive immunity is fully activated.
Figures
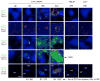
Figure 1. Innate cell clustering and IFNγ production in response to Listeria
(A) Positioning of myeloid (CD11b+), NK cells (NK1.1+) and B cells (B220+) in the spleens from mice either uninfected or infected with Listeria 24 hr previously. T and B areas labeled; RP, red pulp; arrows, central arteriole; 40X. (B) Positioning of IFNγ-producing cells by immunohistochemistry. Conditions as in legend to (A). (C) Gating strategy for NK cells, NKT cells and T cells in spleen before (−) and 24 hr after (+) Listeria infection. Intracellular IFNγ shown in right panels for cell types designated. (D) Percentages of IFNγ-positive cells among designated cell types before (−) and 24 hr after (+) infection with Listeria. n = 3 for (−), 4 for (+); red bars, means. (E) Total numbers of IFNγ-positive cells in the spleen before (−) and 24 hr after (+) infection with Listeria. n = 3 for (−), 4 for (+); red bars, means.
Figure 2. Time course of innate immune response after Listeria infection in spleen
Spleens from the IFNγ-reporter mice infected with Listeria were harvested at the indicated times. Molecules stained for are color-matched and shown to the left of each row: eCFP, enhanced cyan fluorescent protein as a marker for IFNγ, green; cytokines (IFNγ, IL-12), red; Thy1.2 staining marks T cells, blue; NK1.1, NK cells, red; CD11b, CD11b+ myeloid cells, green in the third and fourth rows, blue in the bottom row; Listeria, red; iNOS, inducible nitric oxide synthase, red; HKLM, heat-killed Listeria; LLO-, listeriolysin O-deficient Listeria; RP, red pulp; WP, white pulp; arrowheads, terminal arteriole; arrow, central arteriole; 40X except the penultimate row (100X).
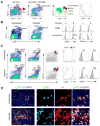
Figure 3. Characterization of CD11b+ myeloid cells in spleen
(A) Gating scheme and markers for the delineation by flow cytometry of plasmacytoid DCs (pDC, orange), conventional DCs (cDC, green), polymorphonuclear leukocytes (PMNs, neutrophils, blue), and monocytes (mono, red) from spleens of uninfected mice. Expression of Ly6G on various myeloid populations which are color-coded as in the third panel and graphed at top right. (B) Simplified scheme to separate neutrophils (PMNs) and monocytes (mono) in uninfected and infected mouse spleen and differentiation of monocytes to TipDCs. Mice were infected 48 hr previously with Listeria. Expression of iNOS and MHC class II from cells gated is depicted, with percentage of monocytes expressing iNOS shown. Gray histogram, isotype control; solid line, iNOS or class II. (C) Distinguishing monocytes and neutrophils by using 7/4, Ly6C and Ly6G. Mice were infected 48 hr previously with Listeria. Percentages of gated cell types among live cells shown. Expression of Ly6G (G1, red line; G2, blue line) and iNOS on gated populations graphed at right. Gray histogram, isotype control; solid line, iNOS. (D) Positioning of monocytes and neutrophils in the spleen of mice before and 24 hr after Listeria infection. The inset areas in the leftmost panels shown at right. WP, white pulp; RP, red pulp; T, T cell area; B, B cell area; arrows, monocytes; arrowheads, neutrophils; 200X.
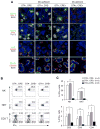
Figure 4. IFNγ from NK cells mediates the maturation of monocytes to TipDCs
(A) Quantitation of numbers of respective cells after depletion and infection with 2000 Listeria shown. n = 6 for each group; **, p<0.01. (B) Immunohistochemistry of mice treated with anti-Gr1 or isotype control antibody and infected 48 hrs later with Listeria. The spleens were examined 24 hr later. Myeloid cell depletion had no effect on NK cell clustering. 40X. (C) Spleen immunohistochemistry of positioning of NK cells, myeloid cells and cells producing IFNγ 24 hr after Listeria infection in wild-type or CCR2-deficient (CCR2 KO) mice. 40X. (D) Mice were treated with anti-asialoGM1 (anti-aGM1) or control Ab (rabbit IgG), and spleens analyzed 48 hr later for proportions of NK cells (uppermost gates) and NKT cells (right gates). Distribution of asialoGM1 staining on NK and NKT cells from spleen denoted in the right panel, with percentage of NKT cells that were stained with anti-aGM1 indicated. (E) NK cell-independent clustering of CD11b+ myeloid cells. Mice were treated with control rabbit antibody (control) or anti-asialoGM1(aGM1) and infected 48 hr later with Listeria. The next day, spleens were harvested and imaged using immunohistochemistry. 40X. (F) Monocytes in the spleen were gated as in Figure 3B and analyzed for expression of iNOS and MHC class II from the wild-type (WT), IFNγ-deficient (IFNγ KO) and RAG1-deficient (RAG KO) mice infected 48 hr earlier with Listeria. Representative flow cytometry plots left, summary of results right. Mean of WT arbitrarily set as 100%. MFI, Mean Fluorescence Intensity; n = 4 for each group; **, p<0.01. (G) Analysis of TipDC-like monocytes in spleen from wild-type (WT) and γc-deficient ( γc KO) mice infected 48 hr earlier with Listeria. Summary with quantitation shown. Mean of WT arbitrarily set as 100%. n = 4 for WT, 3 for γc-deficient group; **, p<0.01. (H) Analysis of TipDC-like monocytes in spleen from RAG1-deficient (RAG KO) mice treated with control rabbit antibody (control) or anti-asialoGM1 antibody (anti-aGM1) and infected with Listeria 48 hr before. Summary with quantitation shown. Mean of control Ab-treated mice arbitrarily set as 100%. n = 3 for control, 4 for anti-aGM1 group; *, p<0.05. (I) Analysis of TipDC-like monocytes in spleen from RAG1-deficient mice and RAG1xIFNγ-deficient (RAG-IFNγ DKO mice infected with Listeria 48 hr before. Summary with quantitation shown. Mean of RAG1-deficient mice arbitrarily set as 100%. n = 3 for each group; **, p<0.01.
Figure 5. CD11c+ cells are required for innate cell clustering and activation in response to Listeria
(A) Spleen immunohistochemistry for indicated markers 24 hr after (upper panels) and before (bottom panels) infection with Listeria. DTA, diphtheria toxin A; Cre, CD11c-cre recombinase; CD169 (sialoadhesin, MOMA-1), a marker for metallophilic macrophages; CD209b (SIGNR1, ER-TR9), a marker for marginal zone macrophages; 40X. (B) Representative flow cytometric analysis of NK cells, NKT cells, and T cells that express intracellular IFNγ when isolated from the indicated mice 24 hr after infection with Listeria. Percentages of IFNγ-positive cells among designated cell types shown. (C) Quantitation of flow cytometric analysis from multiple experiments. *, p<0.05; **, p<0.01.
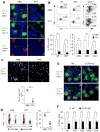
Figure 6. NK cell clustering and activation requires a pertussis-toxin sensitive receptor
(A) Spleen immunohistochemistry for indicated markers 24 hr after Listeria infection of mice pre-treated with PBS or pertussis toxin (PTX). 40X. (B) Representative (upper panels, percentages of IFNγ-positive cells among designated cell types shown) and quantitative summary (lower panels) of intracellular IFNγ detection from the indicated spleen cells 24 hr after Listeria infection. n= 4 for each group; **, p<0.01. (C) NK cells purified from RAG1-deficient mice were labeled with CFSE and treated with PBS (vehicle control) or PTX before transfer to recipient WT mice. Spleen sections were stained for CFSE and IFNγ 24 hr after infection with Listeria. Quantitation of movement of donor (CFSE+), as compared to host (CFSE−) NK cells into clusters, as defined by association with an IFNγ-producing cluster, from multiple experiments is shown below. n = 3 for PBS, 4 for PTX group; **, p<0.01. (D) NK cells prepared as in (C) were isolated from infected mice 24 hr after Listeria infection and stained for IFNγ Loss of IFNγ production in PTX-treated NK cells shown in graph. Mean of IFNγ-producing host cells was set arbitrarily as 100%. n = 4 for each group; **, p<0.01. (E) Spleen immunohistochemistry of positioning of NK cells, myeloid cells, and cells producing IFNγ 24 hr after Listeria infection in wild-type or CCR5-deficient (CCR5 KO) mice. 40X. (F) Quantitation of flow cytometric analysis from multiple experiments of spleen cells staining for intracellular IFNγ 24 hr after Listeria infection from the designated mice. Mean of WT was set arbitrarily as 100%; n = 9 for WT, 8 for CCR5 KO group; *, p<0.05, **, p<0.01.
Figure 7. Signaling pathways involved in innate immune cell clustering and IFNγ production
(A)–(F), Immunohistochemistry of co-infected matched WT or designated knockout (KO) mice to assess NK cell clustering (upper panels) and IFNγ production (lower panels) 24 hr after Listeria infection.
Comment in
- Coordinating innate immune cells to optimize microbial killing.
Serbina NV, Pamer EG. Serbina NV, et al. Immunity. 2008 Nov 14;29(5):672-4. doi: 10.1016/j.immuni.2008.10.003. Immunity. 2008. PMID: 19006691 Free PMC article.
Similar articles
- Sources of interferon-gamma (IFN-gamma) in early immune response to Listeria monocytogenes.
Thäle C, Kiderlen AF. Thäle C, et al. Immunobiology. 2005;210(9):673-83. doi: 10.1016/j.imbio.2005.07.003. Immunobiology. 2005. PMID: 16323704 - Expression of the p60 autolysin enhances NK cell activation and is required for listeria monocytogenes expansion in IFN-gamma-responsive mice.
Humann J, Bjordahl R, Andreasen K, Lenz LL. Humann J, et al. J Immunol. 2007 Feb 15;178(4):2407-14. doi: 10.4049/jimmunol.178.4.2407. J Immunol. 2007. PMID: 17277147 - Activation of naive NK cells in response to Listeria monocytogenes requires IL-18 and contact with infected dendritic cells.
Humann J, Lenz LL. Humann J, et al. J Immunol. 2010 May 1;184(9):5172-8. doi: 10.4049/jimmunol.0903759. Epub 2010 Mar 29. J Immunol. 2010. PMID: 20351186 Free PMC article. - Monocyte-mediated immune defense against murine Listeria monocytogenes infection.
Serbina NV, Shi C, Pamer EG. Serbina NV, et al. Adv Immunol. 2012;113:119-34. doi: 10.1016/B978-0-12-394590-7.00003-8. Adv Immunol. 2012. PMID: 22244581 Free PMC article. Review. - Early events regulating immunity and pathogenesis during Listeria monocytogenes infection.
Williams MA, Schmidt RL, Lenz LL. Williams MA, et al. Trends Immunol. 2012 Oct;33(10):488-95. doi: 10.1016/j.it.2012.04.007. Epub 2012 Jun 5. Trends Immunol. 2012. PMID: 22677184 Free PMC article. Review.
Cited by
- Recognition and Regulation of T Cells by NK Cells.
Pallmer K, Oxenius A. Pallmer K, et al. Front Immunol. 2016 Jun 24;7:251. doi: 10.3389/fimmu.2016.00251. eCollection 2016. Front Immunol. 2016. PMID: 27446081 Free PMC article. Review. - Systems infection biology: a compartmentalized immune network of pig spleen challenged with Haemophilus parasuis.
Zhao M, Liu XD, Li XY, Chen HB, Jin H, Zhou R, Zhu MJ, Zhao SH. Zhao M, et al. BMC Genomics. 2013 Jan 22;14:46. doi: 10.1186/1471-2164-14-46. BMC Genomics. 2013. PMID: 23339624 Free PMC article. - Splenic red pulp macrophages produce type I interferons as early sentinels of malaria infection but are dispensable for control.
Kim CC, Nelson CS, Wilson EB, Hou B, DeFranco AL, DeRisi JL. Kim CC, et al. PLoS One. 2012;7(10):e48126. doi: 10.1371/journal.pone.0048126. Epub 2012 Oct 29. PLoS One. 2012. PMID: 23144737 Free PMC article. - Transient Receptor Potential Melastatin 2 (TRPM2) ion channel is required for innate immunity against Listeria monocytogenes.
Knowles H, Heizer JW, Li Y, Chapman K, Ogden CA, Andreasen K, Shapland E, Kucera G, Mogan J, Humann J, Lenz LL, Morrison AD, Perraud AL. Knowles H, et al. Proc Natl Acad Sci U S A. 2011 Jul 12;108(28):11578-83. doi: 10.1073/pnas.1010678108. Epub 2011 Jun 27. Proc Natl Acad Sci U S A. 2011. PMID: 21709234 Free PMC article. - Memory-T-cell-derived interferon-γ instructs potent innate cell activation for protective immunity.
Soudja SM, Chandrabos C, Yakob E, Veenstra M, Palliser D, Lauvau G. Soudja SM, et al. Immunity. 2014 Jun 19;40(6):974-88. doi: 10.1016/j.immuni.2014.05.005. Epub 2014 Jun 12. Immunity. 2014. PMID: 24931122 Free PMC article.
References
- Aichele P, Zinke J, Grode L, Schwendener RA, Kaufmann SH, Seiler P. Macrophages of the splenic marginal zone are essential for trapping of blood-borne particulate antigen but dispensable for induction of specific T cell responses. J Immunol. 2003;171:1148–1155. - PubMed
- Andrews DM, Farrell HE, Densley EH, Scalzo AA, Shellam GR, Degli-Esposti MA. NK1.1+ cells and murine cytomegalovirus infection: what happens in situ? J Immunol. 2001;166:1796–1802. - PubMed
- Aoshi T, Zinselmeyer BH, Konjufca V, Lynch JN, Zhang X, Koide Y, Miller MJ. Bacterial Entry to the Splenic White Pulp Initiates Antigen Presentation to CD8(+) T Cells. Immunity 2008 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P01 AI078869/AI/NIAID NIH HHS/United States
- R37 AI026918-20/AI/NIAID NIH HHS/United States
- AI026918/AI/NIAID NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- R01 AI026918/AI/NIAID NIH HHS/United States
- AI067804/AI/NIAID NIH HHS/United States
- R37 AI026918/AI/NIAID NIH HHS/United States
- P01 AI078869-019001/AI/NIAID NIH HHS/United States
- R01 AI030663/AI/NIAID NIH HHS/United States
- P01 AI078869-010004/AI/NIAID NIH HHS/United States
- AI078869/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases