Experience-dependent epigenetic modifications in the central nervous system - PubMed (original) (raw)
Review
Experience-dependent epigenetic modifications in the central nervous system
J David Sweatt. Biol Psychiatry. 2009.
Abstract
This mini-review describes recent discoveries demonstrating that experience can drive the production of epigenetic marks in the adult nervous system and that the experience-dependent regulation of epigenetic molecular mechanisms in the mature central nervous system participates in the control of gene transcription underlying the formation of long-term memories. In the mammalian experimental systems investigated thus far, epigenetic mechanisms have been linked to associative fear conditioning, extinction of learned fear, and hippocampus-dependent spatial memory formation. Intriguingly, in one experimental system epigenetic marks at the level of chromatin structure (histone acetylation) have been linked to the recovery of memories that had seemed to be "lost" (i.e., not available for recollection). Environmental enrichment has long been known to have positive effects on memory capacity, and recent studies have suggested that these effects are at least partly due to the recruitment of epigenetic mechanisms by environmental enrichment. Finally, an uncoupling of signal transduction pathways from the regulation of epigenetic mechanisms in the nucleus has been implicated in the closure of developmental critical periods. Taken together, these eclectic findings suggest a new perspective on experience-dependent dynamic regulation of epigenetic mechanisms in the adult nervous system and their relevance to biological psychiatry.
Conflict of interest statement
Disclosure Statement
Dr. Sweatt receives grant funding from the NIH, Evelyn F. McKnight Brain Research Foundation, the Rotary Clubs CART fund, and NARSAD. Dr. Sweatt reports no biomedical financial interests or potential conflicts of interest. The author also wishes to thank Felecia Hester for her assistance in preparing this review.
Figures
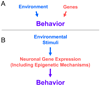
Figure 1
The historical model with separate and distinct influences of genes and environment on behavior (Panel A) is now known to be over-simplified. Instead, contemporary studies have illustrated that environment and experience act in part through altering gene readout in the CNS in order to achieve their effects on behavior (Panel B). One component of the processes by which the environment and experience alter individual behavior includes epigenetic molecular mechanisms such as regulation of chromatin structure and DNA methylation. The historical dichotomy between “nature” (genes) and “nurture” (environment and experience) is too simplistic – genes and experience are mechanistically intertwined. Epigenetic molecular mechanisms contribute to this intertwining.
Figure 2
A simplified scheme for DNA methylation-dependent gene silencing. Methylation of cytosines at CpG dinucleotides (red lollipops) recruits methyl-DNA binding proteins locally to specific sites in the genome. All proteins that bind to methylated DNA have both a methyl-DNA binding domain (MBD) and a transcription-regulatory domain (TRD). The TRD recruits adapter proteins which in turn recruit histone de-acetylases (HDACs). The HDACs alter chromatin structure locally through removing acetyl groups (Ac) from histone core proteins (grey spheres), leading to compaction of chromatin and transcriptional suppression. It is important to note that while this is the traditional and well-established role of methyl-DNA binding proteins in transcriptional regulation, recent findings also support the idea that DNA methylation can also be associated with transcriptional activation.
Similar articles
- Epigenetic modifications in the nervous system and their impact upon cognitive impairments.
Rudenko A, Tsai LH. Rudenko A, et al. Neuropharmacology. 2014 May;80:70-82. doi: 10.1016/j.neuropharm.2014.01.043. Epub 2014 Feb 1. Neuropharmacology. 2014. PMID: 24495398 Review. - Epigenetic mechanisms in memory formation.
Levenson JM, Sweatt JD. Levenson JM, et al. Nat Rev Neurosci. 2005 Feb;6(2):108-18. doi: 10.1038/nrn1604. Nat Rev Neurosci. 2005. PMID: 15654323 Review. - Epigenetic mechanisms in cognition.
Day JJ, Sweatt JD. Day JJ, et al. Neuron. 2011 Jun 9;70(5):813-29. doi: 10.1016/j.neuron.2011.05.019. Neuron. 2011. PMID: 21658577 Free PMC article. Review. - Hippocampus development and function: role of epigenetic factors and implications for cognitive disease.
Lagali PS, Corcoran CP, Picketts DJ. Lagali PS, et al. Clin Genet. 2010 Oct;78(4):321-33. doi: 10.1111/j.1399-0004.2010.01503.x. Clin Genet. 2010. PMID: 20681996 Review. - Epigenetic regulation of memory: implications in human cognitive disorders.
Kramer JM. Kramer JM. Biomol Concepts. 2013 Feb;4(1):1-12. doi: 10.1515/bmc-2012-0026. Biomol Concepts. 2013. PMID: 25436561 Review.
Cited by
- Genomic targets, and histone acetylation and gene expression profiling of neural HDAC inhibition.
Lopez-Atalaya JP, Ito S, Valor LM, Benito E, Barco A. Lopez-Atalaya JP, et al. Nucleic Acids Res. 2013 Sep;41(17):8072-84. doi: 10.1093/nar/gkt590. Epub 2013 Jul 1. Nucleic Acids Res. 2013. PMID: 23821663 Free PMC article. - NF-κB in Innate Neuroprotection and Age-Related Neurodegenerative Diseases.
Lanzillotta A, Porrini V, Bellucci A, Benarese M, Branca C, Parrella E, Spano PF, Pizzi M. Lanzillotta A, et al. Front Neurol. 2015 May 20;6:98. doi: 10.3389/fneur.2015.00098. eCollection 2015. Front Neurol. 2015. PMID: 26042083 Free PMC article. Review. - Benefits of histone deacetylase inhibitors for acute brain injury: a systematic review of animal studies.
Gibson CL, Murphy SP. Gibson CL, et al. J Neurochem. 2010 Nov;115(4):806-13. doi: 10.1111/j.1471-4159.2010.06993.x. Epub 2010 Oct 7. J Neurochem. 2010. PMID: 20831615 Free PMC article. Review. - Regulation of chromatin structure in memory formation.
Roth TL, Sweatt JD. Roth TL, et al. Curr Opin Neurobiol. 2009 Jun;19(3):336-42. doi: 10.1016/j.conb.2009.05.011. Epub 2009 Jun 17. Curr Opin Neurobiol. 2009. PMID: 19539459 Free PMC article. Review. - Histone H3 Acetylation is Asymmetrically Induced Upon Learning in Identified Neurons of the Food Aversion Network in the Mollusk Helix Lucorum.
Danilova AB, Kharchenko OA, Shevchenko KG, Grinkevich LN. Danilova AB, et al. Front Behav Neurosci. 2010 Nov 24;4:180. doi: 10.3389/fnbeh.2010.00180. eCollection 2010. Front Behav Neurosci. 2010. PMID: 21151377 Free PMC article.
References
- Waddington CH. The Strategy of the Genes. New York: MacMillan; 1957.
- Okano M, Xie S, Li E. Cloning and characterization of a family of novel mammalian DNA (cytosine-5) methyltransferases. Nat Genet. 1998;19:219–220. - PubMed
- Chen L, et al. Direct identification of the active-site nucleophile in a DNA (cytosine-5)-methyltransferase. Biochemistry. 1991;30:11018–11025. - PubMed
- Santi DV, Garrett CE, Barr PJ. On the mechanism of inhibition of DNA-cytosine methyltransferases by cytosine analogs. Cell. 1983;33:9–10. - PubMed
- Bird AP. Use of restriction enzymes to study eukaryotic DNA methylation: II. The symmetry of methylated sites supports semi-conservative copying of the methylation pattern. J Mol Biol. 1978;118:49–60. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 AG031722/AG/NIA NIH HHS/United States
- R01 AG031722-34/AG/NIA NIH HHS/United States
- R01 MH057014/MH/NIMH NIH HHS/United States
- R01 MH057014-15/MH/NIMH NIH HHS/United States
LinkOut - more resources
Full Text Sources