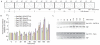A role for the ESCRT system in cell division in archaea - PubMed (original) (raw)
A role for the ESCRT system in cell division in archaea
Rachel Y Samson et al. Science. 2008.
Abstract
Archaea are prokaryotic organisms that lack endomembrane structures. However, a number of hyperthermophilic members of the Kingdom Crenarchaea, including members of the Sulfolobus genus, encode homologs of the eukaryotic endosomal sorting system components Vps4 and ESCRT-III (endosomal sorting complex required for transport-III). We found that Sulfolobus ESCRT-III and Vps4 homologs underwent regulation of their expression during the cell cycle. The proteins interacted and we established the structural basis of this interaction. Furthermore, these proteins specifically localized to the mid-cell during cell division. Overexpression of a catalytically inactive mutant Vps4 in Sulfolobus resulted in the accumulation of enlarged cells, indicative of failed cell division. Thus, the archaeal ESCRT system plays a key role in cell division.
Figures
Fig. 1
(A) Flow cytometric analysis of samples taken at the indicated time points during the progression of a synchronised culture of S. acidocaldarius. The positions of peaks corresponding to one chromosome content (1C) and 2C genome contents are indicated. The cell-cycle stages are indicated at the bottom. (B) Real-time PCR measurements of transcript abundance of the indicated ESCRT-III homologs, Vps4 and a housekeeping control (the nusG transcription elongation factor). Experiments were performed in triplicate, and the standard deviation (SD) is indicated by the error bars. All samples were normalised to the level detected at 0 min. (C) Western blot analysis of the levels of Saci1373 (ESCRT-III), Vps4 and the general transcription factor TATA box binding protein (TBP) proteins across the cell cycle. Samples were taken at the indicated times during the growth of synchronised S. acidocaldarius cultures.
Fig. 2
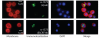
Localisation of (Top) Saci1372 (Vps4) and (bottom) Saci1373 (ESCRT-III). Representative images are shown. Images show the FM4-64X staining for membrane (red), DAPI staining for DNA (blue), antibody labelling of ESCRT-III or Vps4 (green) and merged images. Scale bar, 1 μm. Additional images are shown in figs. S6 and S7 and movie S1.
Fig. 3
(A) Interactions between ESCRT-III proteins detected by yeast 2-hybrid analyses. Yeast containing the indicated plasmids were plated in media lacking leucine and tryptophan to select for plasmids and additionally lacking histidine to score for interactions. (B) Identification of the minimal interaction site on Saci1373 (ESCRT-III) for binding Vps4. Glutathione S-transferase (GST) fusions of ESCRT-III fragments were used in pull-down assays with the full-length Vps4. The results of the pull-downs are shown in the lower panel. The input lanes contain 25 and 5% of input. (C) Identification of the interaction domain of Vps4 that binds Saci1373 (ESCRT-III). GST-Saci1373 (ESCRT-III) was used in pull-down assays with the full-length (1), C-terminal AAA+ domain (2) or isolated MIT domain (3) of Vps4. (D) A schematic representation of the interaction of Saci1373 (red) with the Saci1372 Vps4 MIT domain (yellow). (E) An illustration of the interaction of yeast Vps4 MIT domain (yellow) with the C-terminal MIM1 motif (blue) of the yeast Vps2 ESCRT-III subunit. The MIM1 motif slots between MIT helices α2 and α3 (14). (F) The Saci1373 MIM2 (red)/ Saci1372 MIT (yellow) interaction is closely related in structure with the CHMP6 MIM2 (green, extended)/VPS4A MIT (gray helices) interaction (20).
Fig. 4
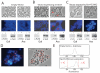
(A to C) Phase contrast and fluorescent microscopy of DAPI stained S. solfataricus cells containing either vector pRYS1 (A), pRYS1-wtVps4 (B) or pRYS1-Vps4 E206Q (C). In each panel, the left-hand images show cells grown in the repressing conditions (in the presence of galactose) and the right-hand images show cells in which expression of the plasmid-encoded gene is induced by the addition of arabinose. Red arrow-heads in (B) indicate enlarged cells with elevated DNA content. The lower panels show the results of western blotting with either antiserum to FLAG or antiserum to Vps4. The antiserum to FLAG detects only the plasmid-encoded Vps4; the antiserum to Vps4 detects both plasmid and chromosomally encoded Vps4. The − and + symbols correspond to cells before and after the addition of either galactose (Gal) or arabinose (Ara). (D) An enlarged image of cells over-expressing Vps4 E206Q (phase contrast image on the left, fluorescent DAPI image on the right). Cells lacking discernable DAPI staining are circled in red. (E) Flow cytometric profile of cells grown in arabinose containing either empty vector or vector over-expressing Walker B (E206Q) Vps4. Cells with less than 1C or more than 2C genome content are indicated.
Similar articles
- Molecular and structural basis of ESCRT-III recruitment to membranes during archaeal cell division.
Samson RY, Obita T, Hodgson B, Shaw MK, Chong PL, Williams RL, Bell SD. Samson RY, et al. Mol Cell. 2011 Jan 21;41(2):186-96. doi: 10.1016/j.molcel.2010.12.018. Mol Cell. 2011. PMID: 21255729 Free PMC article. - Analysis of the Archaeal ESCRT Apparatus.
Samson RY, Duggin IG, Bell SD. Samson RY, et al. Methods Mol Biol. 2019;1998:1-11. doi: 10.1007/978-1-4939-9492-2_1. Methods Mol Biol. 2019. PMID: 31250290 - Coevolution of Eukaryote-like Vps4 and ESCRT-III Subunits in the Asgard Archaea.
Lu Z, Fu T, Li T, Liu Y, Zhang S, Li J, Dai J, Koonin EV, Li G, Chu H, Li M. Lu Z, et al. mBio. 2020 May 19;11(3):e00417-20. doi: 10.1128/mBio.00417-20. mBio. 2020. PMID: 32430468 Free PMC article. - The Structure, Function and Roles of the Archaeal ESCRT Apparatus.
Samson RY, Dobro MJ, Jensen GJ, Bell SD. Samson RY, et al. Subcell Biochem. 2017;84:357-377. doi: 10.1007/978-3-319-53047-5_12. Subcell Biochem. 2017. PMID: 28500532 Review. - Evolution of diverse cell division and vesicle formation systems in Archaea.
Makarova KS, Yutin N, Bell SD, Koonin EV. Makarova KS, et al. Nat Rev Microbiol. 2010 Oct;8(10):731-41. doi: 10.1038/nrmicro2406. Epub 2010 Sep 6. Nat Rev Microbiol. 2010. PMID: 20818414 Free PMC article. Review.
Cited by
- Membrane remodelling in bacteria.
Bohuszewicz O, Liu J, Low HH. Bohuszewicz O, et al. J Struct Biol. 2016 Oct;196(1):3-14. doi: 10.1016/j.jsb.2016.05.010. Epub 2016 Jun 2. J Struct Biol. 2016. PMID: 27265614 Free PMC article. Review. - Overcoming stochastic variations in culture variables to quantify and compare growth curve data.
Sausen CW, Bochman ML. Sausen CW, et al. Bioessays. 2021 Aug;43(8):e2100108. doi: 10.1002/bies.202100108. Epub 2021 Jun 14. Bioessays. 2021. PMID: 34128245 Free PMC article. - The cell biology of archaea.
van Wolferen M, Pulschen AA, Baum B, Gribaldo S, Albers SV. van Wolferen M, et al. Nat Microbiol. 2022 Nov;7(11):1744-1755. doi: 10.1038/s41564-022-01215-8. Epub 2022 Oct 17. Nat Microbiol. 2022. PMID: 36253512 Free PMC article. Review. - An archaeal origin for the actin cytoskeleton: Implications for eukaryogenesis.
Bernander R, Lind AE, Ettema TJ. Bernander R, et al. Commun Integr Biol. 2011 Nov 1;4(6):664-7. doi: 10.4161/cib.16974. Commun Integr Biol. 2011. PMID: 22446522 Free PMC article. - The Calpain-7 protease functions together with the ESCRT-III protein IST1 within the midbody to regulate the timing and completion of abscission.
Paine EL, Skalicky JJ, Whitby FG, Mackay DR, Ullman KS, Hill CP, Sundquist WI. Paine EL, et al. Elife. 2023 Sep 29;12:e84515. doi: 10.7554/eLife.84515. Elife. 2023. PMID: 37772788 Free PMC article.
References
- Karner MB, DeLong EF, Karl DM. Nature. 2001;409:507. - PubMed
- Santelli CM, et al. Nature. 2008;453:653. - PubMed
- Pogliano J. Curr. Op. Cell Biol. 2008;20:19. - PubMed
- Dye NA, Shapiro L. Trends Cell Biol. 2007;17:239. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources