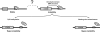CTCF cis-regulates trinucleotide repeat instability in an epigenetic manner: a novel basis for mutational hot spot determination - PubMed (original) (raw)
. 2008 Nov;4(11):e1000257.
doi: 10.1371/journal.pgen.1000257. Epub 2008 Nov 14.
Katharine A Hagerman, Victor V Pineda, Rachel Lau, Diane H Cho, Sandy L Baccam, Michelle M Axford, John D Cleary, James M Moore, Bryce L Sopher, Stephen J Tapscott, Galina N Filippova, Christopher E Pearson, Albert R La Spada
Affiliations
- PMID: 19008940
- PMCID: PMC2573955
- DOI: 10.1371/journal.pgen.1000257
CTCF cis-regulates trinucleotide repeat instability in an epigenetic manner: a novel basis for mutational hot spot determination
Randell T Libby et al. PLoS Genet. 2008 Nov.
Abstract
At least 25 inherited disorders in humans result from microsatellite repeat expansion. Dramatic variation in repeat instability occurs at different disease loci and between different tissues; however, cis-elements and trans-factors regulating the instability process remain undefined. Genomic fragments from the human spinocerebellar ataxia type 7 (SCA7) locus, containing a highly unstable CAG tract, were previously introduced into mice to localize cis-acting "instability elements," and revealed that genomic context is required for repeat instability. The critical instability-inducing region contained binding sites for CTCF -- a regulatory factor implicated in genomic imprinting, chromatin remodeling, and DNA conformation change. To evaluate the role of CTCF in repeat instability, we derived transgenic mice carrying SCA7 genomic fragments with CTCF binding-site mutations. We found that CTCF binding-site mutation promotes triplet repeat instability both in the germ line and in somatic tissues, and that CpG methylation of CTCF binding sites can further destabilize triplet repeat expansions. As CTCF binding sites are associated with a number of highly unstable repeat loci, our findings suggest a novel basis for demarcation and regulation of mutational hot spots and implicate CTCF in the modulation of genetic repeat instability.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
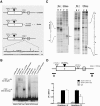
Figure 1. Analysis and mutagenesis of the SCA7-CTCF-I binding site.
(A) SCA7 genomic fragments used for transgenesis. Upper: _SCA7-CTCF-I_-wt; Middle: α-SCA7 3′ genomic deletion; Bottom: _SCA7-CTCF-I_-mut. Core CCCTC sequences are underlined, and sequence alterations in the _SCA7-CTCF-I_-mut transgenic construct are shown in gray. (B) Electrophoretic mobility shift assays with _SCA7-CTCF-I_-wt and -mut probe fragments were performed with probe only, empty lysate (no protein), full-length CTCF protein with pre-immune anti-CTCF sera (CTCF+pI), CTCF protein with anti-CTCF sera (CTCF+α-CTCF), or the 11 zinc-finger DNA binding domain region of CTCF. Arrows indicate shifted CTCF-DNA complexes. Addition of CTCF-DM1 probe as cold competitor prevented CTCF-DNA complex formation for _SCA7-CTCF-I_-wt fragment, while non-specific cold competitor did not (data not shown). (C) Methylation interference (Me I) and DNase I footprinting (DNase) on SCA7-CTCF-I fragment. Left and right panels correspond to the 5′-end labeled coding and anti-sense strands respectively. B, CTCF-bound DNA; F, free DNA; long bars, CTCF-protected from DNase I; arrows, DNase I hypersensitive sites created by CTCF binding; filled circles, contact guanine nucleotides essential for sequence recognition by CTCF. See panel ‘A’ for precise location of sites. (D) ChIP on cerebellar lysates from _SCA7-CTCF-I_-wt and -mut mice (n = 3/genotype). Significantly decreased occupancy at the CTCF-I site was detected with the 3′ amplicon (primer set B) in _SCA7-CTCF-I_-mut mice (p = 0.02, one-way ANOVA), as this amplicon is not in close proximity to the 5′ CTCF-II site. No differences in CTCF occupancy between _SCA7-CTCF-I_-wt and -mut mice were detected with primer set A (or other adjacent primer sets; data not shown) due to the close proximity of the two CTCF binding sites. Results are normalized to _SCA7-CTCF-I_-wt. Error bars are s.d.
Figure 2. _SCA7-CTCF-I_-mut mice display increased germ line instability.
(A) Comparison of CAG repeat instability in parent-offspring transmissions for SCA7-CTCF-I mice. Repeat lengths are plotted as % of total alleles scored for 53 _SCA7-CTCF-I_-wt and 95 _SCA7-CTCF-I_-mut mice. The repeat size range in the _SCA7-CTCF-I_-mut mice was significantly different from the distribution of repeat alleles in the _SCA7-CTCF-I_-wt mice (p = 0.002; Mann-Whitney two-tailed test). (B) Small-pool PCR of sperm DNAs in 16 month-old SCA7 transgenic mice. _SCA7-CTCF-I_-wt mice typically exhibited small repeat length changes, while _SCA7-CTCF-I_-mut mice displayed pronounced instability. (C) Compilation of small-pool PCR data. At 2 months of age, only modest instability was noted. At 16 months of age, _SCA7-CTCF-I_-wt mice displayed moderate instability, but _SCA7-CTCF-I_-mut mice exhibited significantly greater instability (p = 0.009; Mann-Whitney two-tailed test).
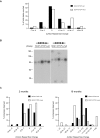
Figure 3. _SCA7-CTCF-I_-mut mice display increased somatic instability.
(A) At 2 months of age, the SCA7 CAG repeat is stable in the _SCA7-CTCF-I_-wt line and in both _SCA7-CTCF-I_-mut lines. (B) With advancing age, tissue-specific instability is seen in _SCA7-CTCF-I_-wt mice; however, this tissue-specific instability is much more pronounced in _SCA7-CTCF-I_-mut mice. Results for individuals from the two different _SCA7-CTCF-I_-mut mice are shown here. (C) To permit quantification of somatic instability, we performed small-pool PCR on tissue DNA samples from _SCA7-CTCF-I_-wt and _SCA7-CTCF-I_-mut mice. As shown here for cortex, _SCA7-CTCF-I_-mut mice displayed significantly greater instability than _SCA7-CTCF-I_-wt mice (p = 8.6×10−5, Mann-Whitney two-tailed test). See Table 1 for a compiled list of repeat alleles. (D) Histogram of repeat length variation in the cortex of _SCA7-CTCF-I_-wt and _SCA7-CTCF-I_-mut mice. _SCA7-CTCF-I_-mut mice exhibit significantly greater instability than _SCA7-CTCF-I_-wt mice, and this expansion tendency exceeds that of _SCA7-CTCF-I_-wt mice, even when 2.5 months younger (p = 0.0003, Mann-Whitney two-tailed test). With advancing age, the expansion bias between the _SCA7-CTCF-I_-mut and -wt mice becomes more pronounced (p<.0001, Mann-Whitney two-tailed test). Results for individuals from the two different _SCA7-CTCF-I_-mut mice are shown here.
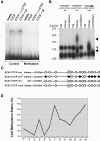
Figure 4. Epigenetic regulation of CTCF binding modulates instability at the SCA7 locus.
(A) CpG methylation prevents binding of CTCF to SCA7-CTCF-I site. Electrophoretic mobility shift assays with un-methylated (control) or methylated SCA7-CTCF-I fragments, using CTCF with no antisera (CTCF), CTCF with anti-CTCF antisera (CTCF+α-CTCF), or CTCF with pre-immune sera (CTCF+pI). Arrow indicates CTCF-bound probe. (B) Prominent somatic instability in kidney DNA (black arrowheads) from a _SCA7-CTCF-I_-wt mouse with CTCF-I site methylation (_SCA7-CTCF-I_-wt*) contrasts with somatic stability in _SCA7-CTCF-I_-wt mice with un-methylated CTCF-I sites. Note that _SCA7-CTCF-I_-wt lines display bimodal CAG repeat alleles. Prominent somatic instability is apparent in kidney DNA (gray arrowhead) from a _SCA7-CTCF-I_-mut mouse. All mice were 6 months of age. (C) Kidney DNAs from the _SCA7-CTCF-I_-wt* mouse are highly methylated. Circles, CpG dyads; open circles, unmethylated; filled circles; methylated. Box highlights core CTCF binding site contact residue, based upon footprinting analysis. Diagrammed epigenotypes summarize results for five _SCA7-CTCF-I_-wt mice, eight _SCA7-CTCF-I_-mut mice, and the _SCA7-CTCF-I_-wt* mouse, and were consistent for at least 75% of all sequenced clones (n = 10−12/sample). (D) Liver DNAs from control _SCA7-CTCF-I_-wt mice are methylated. Bisulfite sequencing of the SCA7-CTCF-I region was performed upon liver DNAs from three _SCA7-CTCF-I_-wt mice at one year of age (n = 17 clones/mouse), and CpG methylation determined for the 13 CpG dyads in the SCA7-CTCF-I region. A number of CpG dyads, including the CpG-4 CTCF contact site, exhibit moderate to high levels of methylation.
Figure 5. Model for CTCF regulation of CAG repeat instability.
Non-expanded CAG repeat is stable, as CTCF is bound to adjacent site. Upon repeat expansion, chromatin environment and DNA structure of repeat region is altered, permitting instability. Loss of CTCF binding at adjacent CTCF binding site, either by CpG methylation or CTCF binding site mutation, further promotes repeat instability.
Similar articles
- A SCA7 CAG/CTG repeat expansion is stable in Drosophila melanogaster despite modulation of genomic context and gene dosage.
Jackson SM, Whitworth AJ, Greene JC, Libby RT, Baccam SL, Pallanck LJ, La Spada AR. Jackson SM, et al. Gene. 2005 Feb 28;347(1):35-41. doi: 10.1016/j.gene.2004.12.008. Gene. 2005. PMID: 15715978 - Genomic context drives SCA7 CAG repeat instability, while expressed SCA7 cDNAs are intergenerationally and somatically stable in transgenic mice.
Libby RT, Monckton DG, Fu YH, Martinez RA, McAbney JP, Lau R, Einum DD, Nichol K, Ware CB, Ptacek LJ, Pearson CE, La Spada AR. Libby RT, et al. Hum Mol Genet. 2003 Jan 1;12(1):41-50. doi: 10.1093/hmg/ddg006. Hum Mol Genet. 2003. PMID: 12490531 - CTCF regulates ataxin-7 expression through promotion of a convergently transcribed, antisense noncoding RNA.
Sopher BL, Ladd PD, Pineda VV, Libby RT, Sunkin SM, Hurley JB, Thienes CP, Gaasterland T, Filippova GN, La Spada AR. Sopher BL, et al. Neuron. 2011 Jun 23;70(6):1071-84. doi: 10.1016/j.neuron.2011.05.027. Neuron. 2011. PMID: 21689595 Free PMC article. - Spinocerebellar ataxia 7 (SCA7).
Lebre AS, Brice A. Lebre AS, et al. Cytogenet Genome Res. 2003;100(1-4):154-63. doi: 10.1159/000072850. Cytogenet Genome Res. 2003. PMID: 14526176 Review. - Molecular pathogenesis and cellular pathology of spinocerebellar ataxia type 7 neurodegeneration.
Garden GA, La Spada AR. Garden GA, et al. Cerebellum. 2008;7(2):138-49. doi: 10.1007/s12311-008-0027-y. Cerebellum. 2008. PMID: 18418675 Free PMC article. Review.
Cited by
- Tissue- and age-specific DNA replication patterns at the CTG/CAG-expanded human myotonic dystrophy type 1 locus.
Cleary JD, Tomé S, López Castel A, Panigrahi GB, Foiry L, Hagerman KA, Sroka H, Chitayat D, Gourdon G, Pearson CE. Cleary JD, et al. Nat Struct Mol Biol. 2010 Sep;17(9):1079-87. doi: 10.1038/nsmb.1876. Epub 2010 Aug 15. Nat Struct Mol Biol. 2010. PMID: 20711191 - Instability and chromatin structure of expanded trinucleotide repeats.
Dion V, Wilson JH. Dion V, et al. Trends Genet. 2009 Jul;25(7):288-97. doi: 10.1016/j.tig.2009.04.007. Epub 2009 Jun 18. Trends Genet. 2009. PMID: 19540013 Free PMC article. Review. - Mechanisms of trinucleotide repeat instability during human development.
McMurray CT. McMurray CT. Nat Rev Genet. 2010 Nov;11(11):786-99. doi: 10.1038/nrg2828. Nat Rev Genet. 2010. PMID: 20953213 Free PMC article. Review. - Histone deacetylase knockouts modify transcription, CAG instability and nuclear pathology in Huntington disease mice.
Kovalenko M, Erdin S, Andrew MA, St Claire J, Shaughnessey M, Hubert L, Neto JL, Stortchevoi A, Fass DM, Mouro Pinto R, Haggarty SJ, Wilson JH, Talkowski ME, Wheeler VC. Kovalenko M, et al. Elife. 2020 Sep 29;9:e55911. doi: 10.7554/eLife.55911. Elife. 2020. PMID: 32990597 Free PMC article. - Phenotypic variance in monozygotic twins with SCA3.
Zhao H, Yang L, Dong Y, Wu ZY. Zhao H, et al. Mol Genet Genomic Med. 2020 Oct;8(10):e1438. doi: 10.1002/mgg3.1438. Epub 2020 Jul 29. Mol Genet Genomic Med. 2020. PMID: 32729243 Free PMC article.
References
- Pearson CE, Nichol Edamura K, Cleary JD. Repeat instability: mechanisms of dynamic mutations. Nat Rev Genet. 2005;6:729–742. - PubMed
- Gouw LG, Castaneda MA, McKenna CK, Digre KB, Pulst SM, et al. Analysis of the dynamic mutation in the SCA7 gene shows marked parental effects on CAG repeat transmission. Hum Mol Genet. 1998;7:525–532. - PubMed
- Monckton DG, Cayuela ML, Gould FK, Brock GJ, Silva R, et al. Very large (CAG)(n) DNA repeat expansions in the sperm of two spinocerebellar ataxia type 7 males. Hum Mol Genet. 1999;8:2473–2478. - PubMed
- La Spada AR, Roling DB, Harding AE, Warner CL, Spiegel R, et al. Meiotic stability and genotype-phenotype correlation of the trinucleotide repeat in X-linked spinal and bulbar muscular atrophy. Nat Genet. 1992;2:301–304. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- EY14061/EY/NEI NIH HHS/United States
- AR050741/AR/NIAMS NIH HHS/United States
- R01 CA068360/CA/NCI NIH HHS/United States
- T32 AG000057/AG/NIA NIH HHS/United States
- AR4203/AR/NIAMS NIH HHS/United States
- AG00057/AG/NIA NIH HHS/United States
- CA68360/CA/NCI NIH HHS/United States
- R01 EY014061/EY/NEI NIH HHS/United States
- R01 GM059356/GM/NIGMS NIH HHS/United States
- GM59356/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials