The Legionella pneumophila replication vacuole: making a cosy niche inside host cells - PubMed (original) (raw)
Review
The Legionella pneumophila replication vacuole: making a cosy niche inside host cells
Ralph R Isberg et al. Nat Rev Microbiol. 2009 Jan.
Abstract
The pathogenesis of Legionella pneumophila is derived from its growth within lung macrophages after aerosols are inhaled from contaminated water sources. Interest in this bacterium stems from its ability to manipulate host cell vesicular-trafficking pathways and establish a membrane-bound replication vacuole, making it a model for intravacuolar pathogens. Establishment of the replication compartment requires a specialized translocation system that transports a large cadre of protein substrates across the vacuolar membrane. These substrates regulate vesicle traffic and survival pathways in the host cell. This Review focuses on the strategies that L. pneumophila uses to establish intracellular growth and evaluates why this microorganism has accumulated an unprecedented number of translocated substrates that are targeted at host cells.
Figures
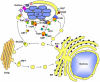
Figure 1. L. pneumophila modulates trafficking of its vacuole to establish a replicative niche
(A) Formation of the replication vacuole. After uptake into target amoebae or macrophages, the “Legionella containing vacuole” (LCV) evades transport to the lysosomal network and is sequestered in a compartment very different from that observed for nonpathogens ,. Within minutes of uptake, vesicles derived from the endoplasmic reticulum (ER; yellow compartments) and mitochondria appear in close proximity to the LCV surface. The identity of the ER-derived vesicles is based on the presence of proteins known to be associated with the early secretory apparatus. The vesicles about the LCV appeared docked and extend out about the surface, and eventually the membranes surrounding the bacterium closely resemble rough ER in appearance, with ribosomes studding them. Within this ER-like compartment, the bacterium replicates to high numbers and eventually lyses the host cell. (B) Default pathway of trafficking nonpathogen. After bacterial uptake, the membrane-bound compartment acquires the character of early endosomes and late endosomes before entering into the lysosomal network.
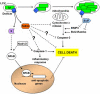
Figure 2
L. pneumophila proteins secreted via the Dot/Icm translocation system associate with the LCV and recruit host proteins involved in vesicle trafficking through the early secretory pathway. For the sole purpose of simplifying the components displayed in the figure, the Dot/Icm apparatus is depicted as a tube extending from the bacterial cytoplasm into the host cytosol, but this there is no mechanistic support of this simplistic view. Sec22b, involved in docking of ER-derived vesicles at the Golgi, is recruited to the LCV, although the mechanism of recruitment is unclear. Rab1, another vesicle docking and fusion protein is recruited to the LCV by the L. pneumophila protein SidM which functions as both a Rab1 GDF (GDI dissociation factor) and a Rab1 GEF (guanine nucleotide exchange factor). LidA acts in conjunction with SidM to sequester activated Rab1 at the LCV membrane. LepB is a RabGAP, and may be involved in dissociation of Rab1 from the vacuolar membrane. Arf1, involved in vesicle budding and recycling at the Golgi, is recruited to the LCV via RalF which functions as an Arf1 GEF. Host membrane recruitment to the LCV may involve an autophagic process as both the host autophagy proteins Atg7 and Atg8 also localize about the LCV.
Figure 3. The Dot/Icm translocation apparatus
Depicted are the presumed locales and topological relationships of the various Dot/Icm components in the L. pneumophila envelope based on a study of the stability of individual proteins in the presence of defined deletion mutations. Individual letters represent Dot protein names whereas letters preceded by an “i” indicated Icm protein names. See text for further details of the individual Dot/Icm components.
Figure 4. L. pneumophila manipulates host cell death and survival pathways
After uptake into mammalian cells there is a response to L. pneumophila that threatens to terminate intracellular growth by causing host cell death. The cell death pathways have both necrotic as well as apoptotic character, and require the presence of an intact Dot/Icm translocation system. The individual L. pneumophila components or translocated substrates that cause cell death have not been identified. In addition, there are at least two translocated substrates that interfere with host cell death. SdhA is required to inhibit multiple pathways that lead to cell death after L. pneumophila contact with host cells, and its absence causes a defect in intracellular replication within macrophages. L. pneumophila also activates the host transcription factor NFκB to promote expression of anti-apoptotic genes to delay host cell death; however, the mechanism by which this occurs has not yet been determined. At later stages of infection, SidF directly inhibits an apoptotic pathway by interfering with pro-death proteins in the Rambo family. See text.
Similar articles
- Immunity to vacuolar pathogens: what can we learn from Legionella?
Neild AL, Roy CR. Neild AL, et al. Cell Microbiol. 2004 Nov;6(11):1011-8. doi: 10.1111/j.1462-5822.2004.00450.x. Cell Microbiol. 2004. PMID: 15469430 Review. - _Legionella_-Containing Vacuoles Capture PtdIns(4)_P_-Rich Vesicles Derived from the Golgi Apparatus.
Weber S, Steiner B, Welin A, Hilbi H. Weber S, et al. mBio. 2018 Dec 11;9(6):e02420-18. doi: 10.1128/mBio.02420-18. mBio. 2018. PMID: 30538188 Free PMC article. - The protein SdhA maintains the integrity of the Legionella-containing vacuole.
Creasey EA, Isberg RR. Creasey EA, et al. Proc Natl Acad Sci U S A. 2012 Feb 28;109(9):3481-6. doi: 10.1073/pnas.1121286109. Epub 2012 Jan 30. Proc Natl Acad Sci U S A. 2012. PMID: 22308473 Free PMC article. - Host Cell S Phase Restricts Legionella pneumophila Intracellular Replication by Destabilizing the Membrane-Bound Replication Compartment.
de Jesús-Díaz DA, Murphy C, Sol A, Dorer M, Isberg RR. de Jesús-Díaz DA, et al. mBio. 2017 Aug 22;8(4):e02345-16. doi: 10.1128/mBio.02345-16. mBio. 2017. PMID: 28830950 Free PMC article. - Phosphoinositide lipids and the Legionella pathogen vacuole.
Haneburger I, Hilbi H. Haneburger I, et al. Curr Top Microbiol Immunol. 2013;376:155-73. doi: 10.1007/82_2013_341. Curr Top Microbiol Immunol. 2013. PMID: 23918172 Review.
Cited by
- Structural and Functional Investigations of the Effector Protein LpiR1 from Legionella pneumophila.
Beyrakhova KA, van Straaten K, Li L, Boniecki MT, Anderson DH, Cygler M. Beyrakhova KA, et al. J Biol Chem. 2016 Jul 22;291(30):15767-77. doi: 10.1074/jbc.M115.708701. Epub 2016 May 17. J Biol Chem. 2016. PMID: 27226543 Free PMC article. - IcmQ in the Type 4b secretion system contains an NAD+ binding domain.
Farelli JD, Gumbart JC, Akey IV, Hempstead A, Amyot W, Head JF, McKnight CJ, Isberg RR, Akey CW. Farelli JD, et al. Structure. 2013 Aug 6;21(8):1361-73. doi: 10.1016/j.str.2013.05.017. Epub 2013 Jul 11. Structure. 2013. PMID: 23850453 Free PMC article. - A pathogen's journey in the host cell: Bridges between actin and traffic.
Franco IS, Shuman HA. Franco IS, et al. Bioarchitecture. 2012 Feb 1;2(2):38-42. doi: 10.4161/bioa.20422. Bioarchitecture. 2012. PMID: 22754628 Free PMC article. - The Legionella IcmSW complex directly interacts with DotL to mediate translocation of adaptor-dependent substrates.
Sutherland MC, Nguyen TL, Tseng V, Vogel JP. Sutherland MC, et al. PLoS Pathog. 2012 Sep;8(9):e1002910. doi: 10.1371/journal.ppat.1002910. Epub 2012 Sep 13. PLoS Pathog. 2012. PMID: 23028312 Free PMC article. - The CRISPR-associated gene cas2 of Legionella pneumophila is required for intracellular infection of amoebae.
Gunderson FF, Cianciotto NP. Gunderson FF, et al. mBio. 2013 Mar 12;4(2):e00074-13. doi: 10.1128/mBio.00074-13. mBio. 2013. PMID: 23481601 Free PMC article.
References
- Muder RR, Yu VL, Woo AH. Mode of transmission of Legionella pneumophila. A critical review. Arch. Intern. Med. 1986;146:1607–12. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources