Noncoding transcription by RNA polymerase Pol IVb/Pol V mediates transcriptional silencing of overlapping and adjacent genes - PubMed (original) (raw)
Noncoding transcription by RNA polymerase Pol IVb/Pol V mediates transcriptional silencing of overlapping and adjacent genes
Andrzej T Wierzbicki et al. Cell. 2008.
Abstract
Nuclear transcription is not restricted to genes but occurs throughout the intergenic and noncoding space of eukaryotic genomes. The functional significance of this widespread noncoding transcription is mostly unknown. We show that Arabidopsis RNA polymerase IVb/Pol V, a multisubunit nuclear enzyme required for siRNA-mediated gene silencing of transposons and other repeats, transcribes intergenic and noncoding sequences, thereby facilitating heterochromatin formation and silencing of overlapping and adjacent genes. Pol IVb/Pol V transcription requires the chromatin-remodeling protein DRD1 but is independent of siRNA biogenesis. However, Pol IVb/Pol V transcription and siRNA production are both required to silence transposons, suggesting that Pol IVb/Pol V generates RNAs or chromatin structures that serve as scaffolds for siRNA-mediated heterochromatin-forming complexes. Pol IVb/Pol V function provides a solution to a paradox of epigenetic control: the need for transcription in order to transcriptionally silence the same region.
Figures
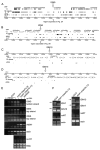
Figure 1. Detection of intergenic Pol V-dependent transcripts
(A–D) Chromosomal contexts of intergenic regions IGN5, IGN6, IGN10 and IGN15. Open reading frames (ORF), transposable element (TE)-derived repeats and small RNAs (sRNA) in the MPSS database (
) are shown. Single copy genes are marked in white, retrotransposons in grey and DNA transposons in black. Diagrams derive from
http://chromatin.cshl.edu/cgi-bin/gbrowse/arabidopsis5/
. (E) Strand-specific RT-PCR analysis of IGN5, IGN6 and AtSN1 transcripts in wild-type (ecotype Col-0), nrpd1a-3, nrpe1 (nrpd1b-11) and nrpd2a-2 nrpd2b-1 mutants. Actin RT-PCR products and ethidium bromide-stained rRNAs resolved by agarose gel electrophoresis serve as loading controls. Dilutions of Col-0 RNA show that PCR results are semi-quantitative. To control for background DNA contamination, a reaction using IGN5 top strand primers, but no reverse transcriptase (no RT) was performed. No RNA (0 μg) controls are provided for all primer pairs. (F) RT-PCR analysis of Pol V-dependent transcripts at intergenic regions _IGN7, IGN10, IGN1_5 and IGN17 in wild-type (Col-0) and nrpe1 mutants.
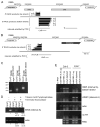
Figure 2. Characterization of Pol V-dependent transcripts
(A, B) Local contexts of IGN5 (A) and IGN6 (B), showing neighboring genes or transposons, 5′ RACE products and intervals amplified by PCR. Color-coding of annotated genes and TE elements is the same as in Fig. 1. For RACE products, the 5′ terminal nucleotide and number of clones (n) sharing that 5′ end are shown. (C) Ethidium bromide-stained agarose gel of 5′ RACE products. (D) 5′ end analysis for Pol V-dependent IGN5 transcripts. RT-PCR was performed on total RNA or RNA treated with Terminator exonuclease or Tobacco Acid Pyrophosphatase. Numbers below the panels are relative densitometric band intensities relative to the untreated control. The mean and standard deviation resulting from three independent experiments is shown. (E) Pol V-dependent transcripts are not polyadenylated. Poly A-enriched and Poly A-depleted RNA fractions were subjected to RT-PCR using IGN5, AtSN1, and actin primer pairs followed by agarose gel electrophoresis and ethidium bromide staining. Controls include no RT (IGN5 bottom strand primers) and no RNA (all primer pairs) reactions.
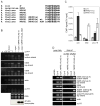
Figure 3. Evidence that Pol V synthesizes IGN transcripts
(A) Multiple alignments of DNA-dependent RNA polymerase largest subunits surrounding the Metal A active site. Invariant aspartates are marked in grey. β′: largest subunit of E. coli polymerase; RPB1: largest subunit of yeast Pol II; NRPA1: largest subunit of Arabidopsis Pol I; NRPB1: largest subunit of Arabidopsis Pol II; NRPC1: largest subunit of Arabidopsis Pol III; NRPD1: largest subunit of Arabidopsis Pol IV (also known as NRPD1a); NRPE1 wt: largest subunit of Arabidopsis Pol V (also known as NRPD1b); NRPE1-ASM: active site mutant of NRPE1. (B) Strand-specific RT-PCR analysis of IGN5 and IGN6 transcripts in Col-0 wild type, nrpe1 (nrpd1b-11), and nrpe1 mutants transformed with a wild-type (wt) FLAG-tagged NRPE1 transgene or the _NRPE1_-ASM transgene. Actin RT-PCR reactions and ethidium bromide-stained rRNAs serve as loading controls. Dilutions of Col-0 wild-type RNA demonstrate that PCR results are semi-quantitative. No RT (IGN5 top strand primers) and no RNA (all primer pairs) controls are included. Equal expression of transgenic wild type and active site mutant NRPE1 was verified by immunoprecipitation followed by αFLAG immunoblot detection (bottom row). (C) ChIP of FLAG-tagged Pol II or Pol V at the actin 2 gene, IGN5 or a solo retroelement long-terminal repeat (LTR) silenced by Pol V. Wild-type Col-0 plants or plants expressing FLAG-tagged NRPB2 or FLAG-tagged NRPE1 were subjected to ChIP using anti-FLAG antibody followed by real-time PCR. Histograms show mean values, +/− standard deviations, obtained for three independent PCR amplifications. (D) RNA immunoprecipitation. Wild-type (non-transgenic) Col-0 and nrpe1 (nrpd1b-11) mutants expressing the NRPE1-FLAG transgene were subjected to RNA-IP using anti-FLAG antibody. Following DNase treatment, IGN5, IGN6, AtSN1, solo LTR or actin 2 RNAs were detected by RT-PCR. AtSN1 and solo LTR PCR-amplified intervals are shown in Figure 4; IGN5 and IGN6 PCR-amplified intervals are shown in Figure 2. Total RNA controls, assayed prior to immunoprecipitation, show that the RNAs are present in equivalent amounts in wild-type Col-0 and NRPE1-FLAG transgenic plants. No RNA and no RT controls used IGN5 top strand primers. No signals were obtained following RNA IP in the absence of anti-FLAG antibody (no AB columns). Background signal for actin RNA shows that equal RNA amounts were tested.
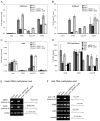
Figure 4. RNA polymerase activity of Pol V is necessary for silencing adjacent transposons and repetitive elements
(A, B) AtSN1 (A) and solo LTR (B) regions, including neighboring genes, repetitive elements and regions amplified by PCR. The diagram for the solo LTR region is based on analysis of transcription units by Huettel et al., 2006. (C) Strand-specific RT-PCR analysis of transcription from the AtSN1 region in Col-0 wild type, nrpe1 (nrpd1b-11) and the nrpe1 mutant expressing a wild-type NRPE1 transgene or the NRPE1-ASM transgene. Intervals amplified by RT-PCR are depicted in panel A. No RT (interval A bottom strand primers) and no RNA controls (all primer pairs) are included. (D) Strand-specific RT-PCR analysis of transcription at the solo LTR region. No RT (interval B bottom strand primers) controls are included. (E) Strand-specific RT-PCR analysis of transcription from a LINE element flanking IGN5. Figure 2A shows the location of interval B amplified by PCR. No RT (interval B bottom strand primers) controls are included. (F) Pol II occupancy of actin 2, IGN5, solo LTR and AtSN1 loci detected using ChIP. Col-0 wild-type, nrpe1 (nrpd1b-11), and nrpe1 mutant plants transformed with the wild type NRPE1 transgene or the NRPE1-ASM transgene were subjected to ChIP using αNRPB2 antibody and detected by real-time PCR. Histograms show the means +/− standard deviations obtained from three independent amplifications.
Figure 5. Pol V-dependent transcription is necessary for heterochromatin formation
(A–C) ChIP using αH3K27me1 (A), αH3K9me2 (B) or αH3Ac (C) antibodies and chromatin of Col-0 wild type, nrpe1 (nrpd1b-11), or nrpe1 mutants transformed with the wild type NRPE1 transgene or NRPE1-ASM transgene. Histograms show the means +/− standard deviations obtained from three independent amplifications. (D) DNA methylation analysis at the indicated loci performed by digestion of genomic DNA with _Mcr_BC followed by quantitative real-time PCR. Comparison to undigested DNA allowed the fraction susceptible to _Mcr_BC to be calculated. (E, F) DNA methylation analysis at the AtSN1, IGN5, IGN6 and solo LTR loci performed by digesting purified DNA with the methylation-sensitive restriction endonucleases _Hae_III (E) or _Alu_I (F) followed by PCR. Sequences lacking _Hae_III (actin; panel E) or _Alu_I (IGN5 interval A; panel F) sites served as controls to show that equivalent amounts of DNA were tested in all reactions.
Figure 6. Pol V-dependent transcription requires the chromatin remodeller, DRD1 but not siRNA production or DNA methylation
(A) Strand-specific RT-PCR analysis of IGN5 and IGN6 transcription in mutants disrupting dicer (dcl1, dcl2, dcl3, dcl4), RNA-dependent RNA polymerase (rdr1, rdr2, rdr6), Pol IV (nrpd1, nrpd2), Pol V (nrpe1/nrpd1b-11, nrpd2) DNA methylation (met1, ddm1, drm2) or chromatin remodelling (ddm1, drd1) activities. Detection of AtSN1 retroelement transcripts indicates a loss of AtSN1 silencing. Col-0 RNA dilutions show that results are semi-quantitative. No RT controls used IGN5 top strand primers. (B) DRD1 is required for Pol V to interact with chromatin. ChIP with αFLAG antibody was performed using chromatin isolated from Col-0 wild-type, nrpe1 (nrpd1b-11) plants expressing the NRPE1-FLAG transgene or drd1 nrpe1 double mutants expressing the NRPE1-FLAG transgene. Actin 2, IGN5, IGN6 and solo LTR loci were detected using quantitative real-time PCR. Histograms show the means +/− standard deviations obtained from three independent amplification reactions. (C) Immunoblot with αFLAG antibody showing that equivalent amounts of NRPE1-FLAG recombinant protein are immunoprecipitated in the nrpe1 (nrpd1b-11) and drd1 nrpe1 genetic backgrounds.
Figure 7
Possible modes of action for Pol V in RNA-directed transcriptional silencing. Pol V transcription and siRNA production occur independently but collaborate in silencing transposons such as AtSN1. 24 nt siRNAs are produced by Pol IV, RDR2 and DCL3 and loaded into AGO4. Chromatin remodeling by DRD1 is required for Pol V to associate with chromatin, and physical interactions may occur between the Pol V C-terminal domain (CTD) and AGO4. In model (A), which we favor, siRNAs bound to AGO4 interact with nascent Pol V transcripts, thereby recruiting chromatin modifying activities, including histone modifying enzymes and the de novo cytosine methyltransferase DRM2, to the adjacent DNA. In (B) AGO4 interacts with the nascent transcripts but the siRNA basepairs with DNA. In (C), the siRNA associated with AGO4 interacts with DNA in a manner dependent upon Pol V-mediated chromatin perturbation.
Comment in
- Pol V transcribes to silence.
Daxinger L, Kanno T, Matzke M. Daxinger L, et al. Cell. 2008 Nov 14;135(4):592-4. doi: 10.1016/j.cell.2008.10.027. Cell. 2008. PMID: 19013268
Similar articles
- Intergenic transcription by RNA polymerase II coordinates Pol IV and Pol V in siRNA-directed transcriptional gene silencing in Arabidopsis.
Zheng B, Wang Z, Li S, Yu B, Liu JY, Chen X. Zheng B, et al. Genes Dev. 2009 Dec 15;23(24):2850-60. doi: 10.1101/gad.1868009. Genes Dev. 2009. PMID: 19948763 Free PMC article. - Metal A and metal B sites of nuclear RNA polymerases Pol IV and Pol V are required for siRNA-dependent DNA methylation and gene silencing.
Haag JR, Pontes O, Pikaard CS. Haag JR, et al. PLoS One. 2009;4(1):e4110. doi: 10.1371/journal.pone.0004110. Epub 2009 Jan 1. PLoS One. 2009. PMID: 19119310 Free PMC article. - RNA polymerase V functions in Arabidopsis interphase heterochromatin organization independently of the 24-nt siRNA-directed DNA methylation pathway.
Pontes O, Costa-Nunes P, Vithayathil P, Pikaard CS. Pontes O, et al. Mol Plant. 2009 Jul;2(4):700-710. doi: 10.1093/mp/ssp006. Epub 2009 Mar 24. Mol Plant. 2009. PMID: 19825650 Free PMC article. - RNA-directed DNA methylation and Pol IVb in Arabidopsis.
Matzke M, Kanno T, Huettel B, Daxinger L, Matzke AJ. Matzke M, et al. Cold Spring Harb Symp Quant Biol. 2006;71:449-59. doi: 10.1101/sqb.2006.71.028. Cold Spring Harb Symp Quant Biol. 2006. PMID: 17381327 Review. - RNA-directed DNA methylation mediated by DRD1 and Pol IVb: a versatile pathway for transcriptional gene silencing in plants.
Huettel B, Kanno T, Daxinger L, Bucher E, van der Winden J, Matzke AJ, Matzke M. Huettel B, et al. Biochim Biophys Acta. 2007 May-Jun;1769(5-6):358-74. doi: 10.1016/j.bbaexp.2007.03.001. Epub 2007 Mar 12. Biochim Biophys Acta. 2007. PMID: 17449119 Review.
Cited by
- Mathematical model of RNA-directed DNA methylation predicts tuning of negative feedback required for stable maintenance.
Dale R, Mosher R. Dale R, et al. Open Biol. 2024 Nov;14(11):240159. doi: 10.1098/rsob.240159. Epub 2024 Nov 13. Open Biol. 2024. PMID: 39532148 Free PMC article. - From RdDM to plant defence.
Martinho C. Martinho C. Nat Plants. 2024 Oct;10(10):1442-1443. doi: 10.1038/s41477-024-01806-9. Nat Plants. 2024. PMID: 39349616 No abstract available. - Epigenetic gene regulation in plants and its potential applications in crop improvement.
Zhang H, Zhu JK. Zhang H, et al. Nat Rev Mol Cell Biol. 2025 Jan;26(1):51-67. doi: 10.1038/s41580-024-00769-1. Epub 2024 Aug 27. Nat Rev Mol Cell Biol. 2025. PMID: 39192154 Review. - An RdDM-independent function of Pol V transcripts in gene regulation and plant defence.
Yuan Y, Liu Y, Han L, Li Y, Qi Y. Yuan Y, et al. Nat Plants. 2024 Oct;10(10):1562-1575. doi: 10.1038/s41477-024-01774-0. Epub 2024 Aug 26. Nat Plants. 2024. PMID: 39187700 - Integrated analysis of DNA methylome and transcriptome revealing epigenetic regulation of CRIR1-promoted cold tolerance.
Li Z, Wang W, Yu X, Zhao P, Li W, Zhang X, Peng M, Li S, Ruan M. Li Z, et al. BMC Plant Biol. 2024 Jul 5;24(1):631. doi: 10.1186/s12870-024-05285-0. BMC Plant Biol. 2024. PMID: 38965467 Free PMC article.
References
- Baulcombe DC. Short silencing RNA: the dark matter of genetics? Cold Spring Harb Symp Quant Biol. 2006;71:13–20. - PubMed
- Brodersen P, Voinnet O. The diversity of RNA silencing pathways in plants. Trends Genet. 2006;22:268–280. - PubMed
- Buhler M, Haas W, Gygi SP, Moazed D. RNAi-dependent and -independent RNA turnover mechanisms contribute to heterochromatic gene silencing. Cell. 2007;129:707–721. - PubMed
- Buhler M, Verdel A, Moazed D. Tethering RITS to a nascent transcript initiates RNAi- and heterochromatin-dependent gene silencing. Cell. 2006;125:873–886. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases