The three fungal transmembrane nuclear pore complex proteins of Aspergillus nidulans are dispensable in the presence of an intact An-Nup84-120 complex - PubMed (original) (raw)
The three fungal transmembrane nuclear pore complex proteins of Aspergillus nidulans are dispensable in the presence of an intact An-Nup84-120 complex
Hui-Lin Liu et al. Mol Biol Cell. 2009 Jan.
Abstract
In Aspergillus nidulans nuclear pore complexes (NPCs) undergo partial mitotic disassembly such that 12 NPC proteins (Nups) form a core structure anchored across the nuclear envelope (NE). To investigate how the NPC core is maintained, we affinity purified the major core An-Nup84-120 complex and identified two new fungal Nups, An-Nup37 and An-ELYS, previously thought to be vertebrate specific. During mitosis the An-Nup84-120 complex locates to the NE and spindle pole bodies but, unlike vertebrate cells, does not concentrate at kinetochores. We find that mutants lacking individual An-Nup84-120 components are sensitive to the membrane destabilizer benzyl alcohol (BA) and high temperature. Although such mutants display no defects in mitotic spindle formation, they undergo mitotic specific disassembly of the NPC core and transient aggregation of the mitotic NE, suggesting the An-Nup84-120 complex might function with membrane. Supporting this, we show cells devoid of all known fungal transmembrane Nups (An-Ndc1, An-Pom152, and An-Pom34) are viable but that An-ndc1 deletion combined with deletion of individual An-Nup84-120 components is either lethal or causes sensitivity to treatments expected to destabilize membrane. Therefore, the An-Nup84-120 complex performs roles, perhaps at the NPC membrane as proposed previously, that become essential without the An-Ndc1 transmembrane Nup.
Figures
Figure 1.
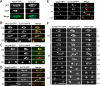
Identification of two novel proteins of the An-Nup84-120 complex. (A) Endogenously S-Tagged SonBcNup96 was affinity purified and resolved using SDS-PAGE. Proteins within the major bands visualized by Coomassie Blue staining were identified using mass spectrometric analysis. Protein identifications are as indicated. d, degradation products. (B) Size and domains of An-Nup37 and An-ELYS are indicated. A WD40-like domain was found in An-Nup37 (see text). (C) Homologues of An-Nup37 and An-ELYS are conserved in filamentous fungi but are absent in S. cerevisiae. n.d., not detected. *, Mans et al. (2004) detected similarity between vertebrate Nup37 and S. pombe SPAC4F10.18. $, predicted gene structure of Afu2g02160 is incorrect. Our predicted length of the An-ELYS homologue in A. fumigatus is indicated. (D) PSI-BLAST analysis indicates that full-length An-ELYS is similar to part of mammalian ELYS (5E-94, ENSP 0000354780, amino acids 681–988), but not to C. elegans MEL-28. The figure is modeled after Galy et al. (2006).
Figure 2.
An-Nup37 and An-ELYS copurify core An-Nup84-120 complex components. (A) Endogenously S-Tagged nucleoporins, as indicated, were affinity purified. Protein extracts of wild-type strain R153, which does not express an S-Tagged protein, were similarly treated to demonstrate specificity of the purifications. Purified proteins were resolved by SDS-PAGE and the gels stained with Coomassie Blue. Bands 1–11 were identified using mass spectroscopy. Band 1, an-Nup120; band 2, an-Nup85; band 3, SonBcNup96; band 4, degradation product of An-Nup120; band 5, an-Nup37; band 6, an-Sec13; band 7, an-ELYS; band 8, an-Nup133; band 9, an-Nup84; and bands 10 and 11, degradation product of An-Nup84. Percent mass spectrometric sequence coverage is detailed in Table 1. S-Tagged proteins are labeled with asterisk (*). (B) An-Nup84-120 components are present in An-Nup37-S-Tag pull downs from an _An-nup133_-deleted strain but not an _An-nup120_-deleted strain. An-Nup84-120 components are not present in an An-ELYS-S-Tag pull down from an _An-nup120_-deleted strain.
Figure 3.
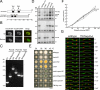
An-Nup37 and An-ELYS are stable NPC core components and are also present on SPBs during mitosis. (A) Endogenously GFP-Tagged An-Nup37 and An-ELYS cells were fixed and stained with DAPI to visualize DNA. A single nonconfocal image is shown. (B–F) The location of endogenously GFP-Tagged Nups as well as SPB and kinetochore markers were followed using live cell spinning disk confocal microscopy. (B) An-ELYS and An-Nup120 colocalize at the NPCs during mitosis. See Supplemental Video 1. (C) An-ELYS remains at NPCs during mitosis when peripheral Nups such as An-Nup49 are dispersed (Supplemental Video 2). (D and E) During mitosis, An-ELYS does not locate to the kinetochores (An-Ndc80) but accumulates at the SPBs (An-Gcp3), which are indicated by arrowheads (Supplemental Video 3). (F) SonBcNup96, An-Nup37, and An-Ndc1 (Supplemental Videos 5–7) remain at the NPCs throughout the cell cycle and also transiently locate to the SPBs during mitosis (indicated by arrowheads). Bar, 5 μm.
Figure 4.
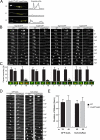
The An-Nup84-120 complex disassembles from NPCs during mitosis in ΔAn-nup37_ΔAn_-elys cells at permissive temperature. (A) Endogenously GFP-Tagged SonBcNup96 remains at the NE during wild-type mitosis (Figure 3F) but is reversibly dispersed from the NE during mitosis in a ΔAn_-nup37_ΔAn_-elys_ background (Supplemental Video 8) at 25°C. The dispersal of SonBcNup96 can be seen in the micrographs and is reflected in the pixel profiles through the G2, M, and G1 nucleus. (B) Similarly, as shown, endogenously GFP-tagged An-Nup120, An-Nup85, An-Nup133 (see wild type [Supplemental Video 9] and ΔAn-nup37_ΔAn-elys [Supplemental Video 10] for comparison), and An-Nup84 remain at the NE during mitosis in wild-type cells but mitotically disperse in a ΔAn_-nup37_ΔAn_-elys double-mutant background. (C) Graphs showing nuclear levels of the Nups from the strains indicated in B during interphase (I) or mitosis (M). The remaining GFP-tagged Nup (green) at the NE was quantified by measuring the net fluorescence intensity of interphase and mitotic nuclei defined by DAPI staining (red). For mitotic samples, cells were treated with 2.4 μg/ml benomyl to arrest cells in a pseudometaphase state. (D and E) GFP-tubA and NLS-DsRed were followed to compare spindle morphology and nuclear transport in wild-type and ΔAn-nup37_ΔAn-elys cells. The duration of mitosis was measured and plotted in the bar graph shown (E). The time difference between wild-type and ΔAn_-nup37_ΔAn_-elys cells is significant (p < 0.05 for tubA-GFP and p < 0.01 for NLS-DsRed). Data are represented as mean ± SD. Bar, 5 μm.
Figure 5.
Locations of core NPC components are modified in ΔAn_-nup37_ΔAn_-elys_ cells. (A–F) Endogenously GFP-tagged An-Nup2, An-Pom152, An-Ndc1, An-Gle1, An-Erg24, and An-Nup170 were analyzed in both wild-type and ΔAn_-nup37_ΔAn_-elys_ cells at 25°C. (A–C) The distribution of An-Nup2 (Supplemental Videos 20 and 21), An-Pom152 (Supplemental Videos 22 and 23), and An-Ndc1 (Supplemental Videos 7 and 24) are similar in wild-type and ΔAn_-nup37_ΔAn_-elys_ cells. (D) An-Gle1 reveals two restrictions of the NE, which generates two nuclei during wild-type (Supplemental Video 25) mitosis (Ukil, De Souza, Liu, and Osmani, unpublished data) indicated by two arrowheads. In ΔAn_-nup37_ΔAn_-elys_ cells (Supplemental Video 26), the double restrictions were not detected but instead a transient bright focus (indicated by an arrow) was observed. (E) An-Erg24-GFP also reveals the double NE restriction in wild-type mitosis (indicated by two arrowheads, Supplemental Video 27), but in ΔAn_-nup37_ΔAn_-elys_ mitotic cells a bright focus accumulates (indicated by an arrow, Supplemental Video 28). (F) An-Nup170 distribution becomes more punctate during mitosis in ΔAn_-nup37_ΔAn_-elys_ cells (Supplemental Video 30) compared with the wild-type (Video 29) mitotic distribution. Bar, 5 μm.
Figure 6.
Interactions between An-Nup84-120 complex components and transmembrane Nups. (A) Cells with the indicated genetic backgrounds were spot inoculated on MAG plates and incubated at 32, 36, and 38°C for 40 h. The white, green, or yellow color of the colonies is caused by different spore (conidia) colors of the strains, which does not affect growth. Bar, 1 cm. The double ΔAn-nup84_ΔAn-pom152 mutations cause synthetic growth defects at all temperatures but can be propagated on 1 M sucrose plates (data not shown). (B) Heterokaryon rescue analysis. After deletion of either An-nup84, An-nup120, or An-nup133 in the ΔAn-ndc1 background allowed recovery of the double mutants only through heterokaryon rescue (Osmani et al., 2006b). Conidia (asexual uninucleated spores) from the heterokaryons were streaked on selective (−UU) and nonselective (+UU) plates and incubated at 32°C for 24 h before photography. Results of diagnostic PCR of wild-type cells, heterokaryotic cells grown on selective media, and spores grown from the heterokaryon on nonselective media are shown, as indicated in the cartoon. The combination of the growth tests and diagnostic PCR results proves (Osmani et al., 2006b) that deletion of An-ndc1 in combination with either ΔAn-nup84, ΔAn-nup120, or ΔAn-nup133 alleles causes lethality. (C) Spores from strains with null alleles of the indicated nonessential NPC core components were spread on MAG or 0.15% BA/MAG plates to test their sensitivity to BA and temperature. Cells on MAG plates were incubated at 36 and 42°C for 48 h. Cells on 0.15% BA/MAG plates were incubated at 36°C for 70 h. Representative colonies are shown for each strain. Bar, 1 cm. (D) Quantification of the tolerance to 0.15% BA at 36°C. Well-separated colonies were chosen for measurement. Colony diameters were measured and plotted in the bar graph. Data are represented as mean ± SD. (E) Representative location of endogenously GFP-tagged An-Nup37 in various genetic backgrounds of cells progressing through mitosis (Supplemental Videos 31–34). Bar, 5 μm. (F) The net intensity of An-Nup37-GFP (green) around interphase and mitotic nuclei defined by DAPI staining (red) in the strains indicated in E, quantified as in Figure 4C. The difference between ΔAn_-elys and ΔAn_-ndc1_ΔAn_-elys_ is significant (p < 0.0001). Data are represented as mean ± SD.
Figure 7.
Triple transmembrane Nup-deleted cells are viable. (A) Size and domains of An-Pom34 and Sc-Pom34 are indicated. Two transmembrane domains predicted using the TMPRED software locate at comparable positions (Miao et al., 2006). (B) Endogenously GFP-tagged An-Pom34 locates around the nuclear periphery similar to An-Nup49 in interphase. An-Pom34 is part of the mitotic NPC core as, unlike the peripheral Nup An-Nup49, it does not disperse during mitotic arrest imposed by SAC activation. (C and D) Both diagnostic PCR and Southern blot analysis were used to confirm deletion of all three transmembrane Nup genes in the same strain. External primers were used to confirm the loci of An_-pom34_, An_-pom152_, and An_-ndc1_ genes have been replaced by the deletion cassettes as indicated by the predicted size differences between bands in the wild-type and triple deletion strain. Southern blot analysis using the coding regions of An_-pom34_, An_-pom152_, An_-histone H1_ and An_-ndc1_ as probes was performed to confirm that none of the known fungal transmembrane Nups are present in triple deleted cells. All four probes as indicated were used (top), whereas the indicated two probes were used (middle and bottom) to distinguish An-ndc1 and An-pom34 deletions whose bands run at near identical size. (E) Cells with the indicated genetic backgrounds were spot inoculated on MAGUU plates and incubated at 20°C for 5 d and at 32, 36, and 42°C for 39 h. Bar, 1 cm. (F) Wild-type and transmembrane Nup-deleted cells were spot inoculated on MAGUU plates at 36°C to follow their growth curve (1, wild type; 2, ΔAn_-pom34_; 3, ΔAn_-ndc1_; 4, ΔAn_-pom152_; 5, ΔAn_-ndc1_ΔAn_-pom152_; 6, ΔAn_-pom34_ΔAn_-pom152_; 7, ΔAn_-ndc1_ΔAn_-pom34_; and 8, ΔAn_-ndc1_ΔAn_-pom34_ΔAn_-pom152_). The diameter of cells was measured every 24 h for 5 d. (G) Spindle morphology (GFP-tubA) and the nuclear transport (NLS-DsRed) of wild type (Supplemental Video 35), and triple transmembrane Nup-deleted (Supplemental Video 36) cells were followed using live cell spinning disk confocal microscopy at 25°C.
Figure 8.
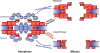
Extreme mitotic changes in the composition of the A. nidulans NPC core structure in ΔAn_-nup37_ΔAn_-elys_ cells. Two new nucleoporins, An-Nup37 and An-ELYS, have been identified as components of the An-Nup84-Nup120 complex of A. nidulans. These nucleoporins are lettered in yellow within the overall proposed structure of the NPC based upon our data in A. nidulans and the extensive studies of others. Both An-Nup37 and An-ELYS are core NPC components, along with An-Nup85, An-Nup120, An-Nup84, An-Nup133, SonBcNup96, An-Pom152, An-Ndc1, An-Pom34, An-Gle1, and An-Nup170 (red), which all remain at NPCs during interphase and mitosis. The peripheral Nups, including SonAGle2, SonBnNup98, An-Nup42, An-Nup49, An-Nup57, An-Nup82, An-Nup159, An-Nup188, An-Nup192, An-Nic96, An-Nsp1, An-Mlp1, An-Sac3, and An-Nup2 (blue), disassemble from NPCs during mitosis. In the absence of An-Nup37 and An-ELYS, the basic composition of the NPC is not changed during interphase. However, during mitosis the entire An-Nup84-120 complex is dispersed from the NPC, demonstrating normal composition of the An-Nup84-120 complex is required to maintain the core NPC structure intact during mitosis. Surprisingly, NPCs can be reassembled normally without the prepore structure when the An-Nup84-120 complex is mitotically dispersed. Although the transmembrane Nups An-Pom152, An-Pom34, and An-Ndc1 as well as An-Nup170 and An-Gle1 are not dispersed in this condition, they are unlikely to form a prepore structure in the NE throughout mitosis and their interactions with each other remains to be determined. However, that An-Pom152, An-Pom34, An-Ndc1, An-Nup170, and An-Gle1 do not disperse during mitosis indicates that they are positioned to play an early role in the NPC reassembly in A. nidulans.
Similar articles
- The mode of mitosis is dramatically modified by deletion of a single nuclear pore complex gene in Aspergillus nidulans.
Chemudupati M, Johns M, Osmani SA. Chemudupati M, et al. Fungal Genet Biol. 2019 Sep;130:72-81. doi: 10.1016/j.fgb.2019.04.010. Epub 2019 Apr 23. Fungal Genet Biol. 2019. PMID: 31026588 - Systematic deletion and mitotic localization of the nuclear pore complex proteins of Aspergillus nidulans.
Osmani AH, Davies J, Liu HL, Nile A, Osmani SA. Osmani AH, et al. Mol Biol Cell. 2006 Dec;17(12):4946-61. doi: 10.1091/mbc.e06-07-0657. Epub 2006 Sep 20. Mol Biol Cell. 2006. PMID: 16987955 Free PMC article. - Nup2 requires a highly divergent partner, NupA, to fulfill functions at nuclear pore complexes and the mitotic chromatin region.
Markossian S, Suresh S, Osmani AH, Osmani SA. Markossian S, et al. Mol Biol Cell. 2015 Feb 15;26(4):605-21. doi: 10.1091/mbc.E14-09-1359. Epub 2014 Dec 24. Mol Biol Cell. 2015. PMID: 25540430 Free PMC article. - Nuclear envelope insertion of spindle pole bodies and nuclear pore complexes.
Jaspersen SL, Ghosh S. Jaspersen SL, et al. Nucleus. 2012 May-Jun;3(3):226-36. doi: 10.4161/nucl.20148. Epub 2012 May 1. Nucleus. 2012. PMID: 22572959 Free PMC article. Review. - Poring over chromosomes: mitotic nuclear pore complex segregation.
Suresh S, Osmani SA. Suresh S, et al. Curr Opin Cell Biol. 2019 Jun;58:42-49. doi: 10.1016/j.ceb.2019.01.002. Epub 2019 Feb 22. Curr Opin Cell Biol. 2019. PMID: 30798206 Review.
Cited by
- Advances in the understanding of nuclear pore complexes in human diseases.
Li Y, Zhu J, Zhai F, Kong L, Li H, Jin X. Li Y, et al. J Cancer Res Clin Oncol. 2024 Jul 30;150(7):374. doi: 10.1007/s00432-024-05881-5. J Cancer Res Clin Oncol. 2024. PMID: 39080077 Free PMC article. Review. - Structure-function mapping of a heptameric module in the nuclear pore complex.
Fernandez-Martinez J, Phillips J, Sekedat MD, Diaz-Avalos R, Velazquez-Muriel J, Franke JD, Williams R, Stokes DL, Chait BT, Sali A, Rout MP. Fernandez-Martinez J, et al. J Cell Biol. 2012 Feb 20;196(4):419-34. doi: 10.1083/jcb.201109008. Epub 2012 Feb 13. J Cell Biol. 2012. PMID: 22331846 Free PMC article. - Comparative genomic evidence for a complete nuclear pore complex in the last eukaryotic common ancestor.
Neumann N, Lundin D, Poole AM. Neumann N, et al. PLoS One. 2010 Oct 8;5(10):e13241. doi: 10.1371/journal.pone.0013241. PLoS One. 2010. PMID: 20949036 Free PMC article. - The Nuclear Pore Complex: Birth, Life, and Death of a Cellular Behemoth.
Dultz E, Wojtynek M, Medalia O, Onischenko E. Dultz E, et al. Cells. 2022 Apr 25;11(9):1456. doi: 10.3390/cells11091456. Cells. 2022. PMID: 35563762 Free PMC article. Review. - Regulated inactivation of the spindle assembly checkpoint without functional mitotic spindles.
De Souza CP, Hashmi SB, Yang X, Osmani SA. De Souza CP, et al. EMBO J. 2011 Jun 3;30(13):2648-61. doi: 10.1038/emboj.2011.176. EMBO J. 2011. PMID: 21642954 Free PMC article.
References
- Alber F., et al. Determining the architectures of macromolecular assemblies. Nature. 2007a;450:683–694. - PubMed
- Alber F., et al. The molecular architecture of the nuclear pore complex. Nature. 2007b;450:695–701. - PubMed
- Antonin W., Ellenberg J., Dultz E. Nuclear pore complex assembly through the cell cycle: regulation and membrane organization. FEBS Lett. 2008;582:2004–2016. - PubMed
- Antonin W., Mattaj I. W. Nuclear pore complexes: round the bend? Nat. Cell Biol. 2005;7:10–12. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases