Unique hexosaminidase reduces metabolic survival signal and sensitizes cardiac myocytes to hypoxia/reoxygenation injury - PubMed (original) (raw)
Unique hexosaminidase reduces metabolic survival signal and sensitizes cardiac myocytes to hypoxia/reoxygenation injury
Gladys A Ngoh et al. Circ Res. 2009.
Abstract
Metabolic signaling through the posttranslational linkage of N-acetylglucosamine (O-GlcNAc) to cellular proteins represents a unique signaling paradigm operative during lethal cellular stress and a pathway that we and others have recently shown to exert cytoprotective effects in vitro and in vivo. Accordingly, the present work addresses the contribution of the hexosaminidase responsible for removing O-GlcNAc (ie, O-GlcNAcase) from proteins. We used pharmacological inhibition, viral overexpression, and RNA interference of O-GlcNAcase in isolated cardiac myocytes to establish its role during acute hypoxia/reoxygenation. Elevated O-GlcNAcase expression significantly reduced O-GlcNAc levels and augmented posthypoxic cell death. Conversely, short interfering RNA directed against, or pharmacological inhibition of, O-GlcNAcase significantly augmented O-GlcNAc levels and reduced posthypoxic cell death. On the mechanistic front, we evaluated posthypoxic mitochondrial membrane potential and found that repression of O-GlcNAcase activity improves, whereas augmentation impairs, mitochondrial membrane potential recovery. Similar beneficial effects on posthypoxic calcium overload were also evident. Such changes were evident without significant alteration in expression of the major putative components of the mitochondrial permeability transition pore (ie, voltage-dependent anion channel, adenine nucleotide translocase, cyclophilin D). The present results provide definitive evidence that O-GlcNAcase antagonizes posthypoxic cardiac myocyte survival. Moreover, such results support a renewed approach to the contribution of metabolism and metabolic signaling to the determination of cell fate.
Conflict of interest statement
Disclosures: None.
Figures
Figure 1
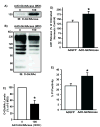
Myocytes (n>/=5/group) were infected with AdGFP or Ad_O_-GlcNAcase (0 or 100 MOI) 48 hours prior to protein isolation or hypoxia-reoxygenation. A) Representative immunoblot of _O_-GlcNAcase protein shows significant elevation in _O_-GlcNAcase levels following Ad_O_-GlcNAcase infection. Representative immunoblot (B) and Densitometric analysis (C) of _O_-GlcNAc levels. Ad_O_-GlcNAcase significantly reduced _O_-GlcNAc levels. As expected, multiple immunopositive bands appear because the _O_-GlcNAc modification occurs on numerous proteins throughout the cell. D) _O_-GlcNAcase overexpression exacerbated post-hypoxic cardiac myocyte damage according to LDH release. E) _O_-GlcNAcase overexpression aggravated post-hypoxic injury according to propidium iodide positivity (n=4/group). *p<0.05 vs. 0 MOI Ad_O_-GlcNAcase or 100 MOI AdGFP.
Figure 2
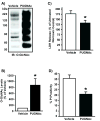
NRCMs were subjected to pharmacologic repression of O-GlcNAcase activity. A) Representative immunoblots for _O_-GlcNAc levels following PUGNAc treatment (n=6/group) show a significant increase in _O_-GlcNAc levels compared to Vehicle. Multiple bands occur because _O_-GlcNAc is a post-translational modification. B) Densitometric analyses of _O_-GlcNAc western blots show significantly elevated _O_-GlcNAc levels compared to Vehicle. C) _O_-GlcNAcase inhibition with PUGNAc diminished post-hypoxic injury in NRCMs (according to LDH release) compared with Vehicle. D) _O_-GlcNAcase inhibition with PUGNAc reduced post-hypoxic injury (per PI positivity) compared with Vehicle. *p<0.05 vs. Vehicle.
Figure 3
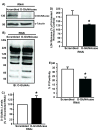
A) _O_-GlcNAcase message knockdown (RNAi) significantly reduced _O_-GlcNAcase protein levels compared with Scr RNAi. B) Representative immunoblot for lysates from Scr vs. _O_-GlcNAcase RNAi NRCMs showing augmented _O_-GlcNAc levels compared to Scr. C) Densitometric analysis of _O_-GlcNAc immunoblots showed significant increase in _O_-GlcNAc levels for _O_-GlcNAcase RNAi compared with Scr RNAi. _O_-GlcNAcase RNAi-treated NRCMs (n=6/group) were more resistant to hypoxia-induced injury according to LDH release (D) and, PI positivity (E) compared to Scr RNAi. *p< 0.05 vs. Scr RNAi.
Figure 4
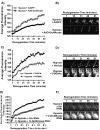
Assessment of sensitivity to loss of mitochondrial membrane potential in NRCMs overexpressing _O_-GlcNAcase (Ad_O_-GlcNAcase) or with inhibition of _O_-GlcNAcase (PUGNAc) following hypoxia. TMRM fluorescence was used to indicate mitochondrial membrane potential (n>/=6/group). A) In post-hypoxic myocytes, _O_-GlcNAcase overexpression (Ad_O_-GlcNAcase) exacerbated mitochondrial membrane potential loss. B) Quantification of the average relative fluorescence intensity for AdGFP and _O_-GlcNAcase overexpression (Ad_O_-GlcNAcase) showed significantly reduced recovery of mitochondrial membrane potential for Ad_O_-GlcNAcase-treated compared to AdGFP-treated cells. C) _O_-GlcNAcase inhibition (PUGNAc) attenuated the loss of mitochondrial membrane potential compared to Vehicle. D) Quantification of the average relative fluorescence intensity for Vehicle and _O_-GlcNAcase inhibition (PUGNAc) showed significant recovery of mitochondrial membrane potential for _O_-GlcNAcase inhibitor (PUGNAc) compared to Vehicle. Similarly, RNAi against _O_-GlcNAcase improved post-hypoxic mitochondrial membrane potential recovery (E&F).
Figure 5
Expression of putative molecular components of mitochondrial permeability transition pore (mPTP). Expression of CypD (A), ANT (B), and VDAC (C) were not significantly affected by genetic overexpression, pharmacologic inhibition, or RNAi.
Figure 6
Evaluation of calcium overload in post-hypoxic cardiac myocytes using Rhod-2AM ((n>/=6/group). Myocytes were treated with AdGFP or Ad_O_-GlcNAcase (A&B), or, Vehicle or PUGNAc (C&D). Following hypoxia myocytes undergo progressive calcium overload. Genetic overexpression of _O_-GlcNAcase exaggerates, while pharmacologic inhibition of _O_-GlcNAcase attenuates post-hypoxic calcium overload.
Comment in
- O-linked beta-N-acetylglucosamine: a new piece of the cardioprotection puzzle?
Downey JM, Cohen MV. Downey JM, et al. Circ Res. 2009 Jan 2;104(1):7-8. doi: 10.1161/CIRCRESAHA.108.191163. Circ Res. 2009. PMID: 19118281 Free PMC article. No abstract available.
Similar articles
- O-GlcNAc signaling attenuates ER stress-induced cardiomyocyte death.
Ngoh GA, Hamid T, Prabhu SD, Jones SP. Ngoh GA, et al. Am J Physiol Heart Circ Physiol. 2009 Nov;297(5):H1711-9. doi: 10.1152/ajpheart.00553.2009. Epub 2009 Sep 4. Am J Physiol Heart Circ Physiol. 2009. PMID: 19734355 Free PMC article. - The role of protein O-linked beta-N-acetylglucosamine in mediating cardiac stress responses.
Chatham JC, Marchase RB. Chatham JC, et al. Biochim Biophys Acta. 2010 Feb;1800(2):57-66. doi: 10.1016/j.bbagen.2009.07.004. Epub 2009 Jul 14. Biochim Biophys Acta. 2010. PMID: 19607882 Free PMC article. Review. - O-linked beta-N-acetylglucosamine: a new piece of the cardioprotection puzzle?
Downey JM, Cohen MV. Downey JM, et al. Circ Res. 2009 Jan 2;104(1):7-8. doi: 10.1161/CIRCRESAHA.108.191163. Circ Res. 2009. PMID: 19118281 Free PMC article. No abstract available. - Non-canonical glycosyltransferase modulates post-hypoxic cardiac myocyte death and mitochondrial permeability transition.
Ngoh GA, Watson LJ, Facundo HT, Dillmann W, Jones SP. Ngoh GA, et al. J Mol Cell Cardiol. 2008 Aug;45(2):313-25. doi: 10.1016/j.yjmcc.2008.04.009. Epub 2008 May 2. J Mol Cell Cardiol. 2008. PMID: 18539296 Free PMC article. - The Role of O-GlcNAcylation for Protection against Ischemia-Reperfusion Injury.
Jensen RV, Andreadou I, Hausenloy DJ, Bøtker HE. Jensen RV, et al. Int J Mol Sci. 2019 Jan 18;20(2):404. doi: 10.3390/ijms20020404. Int J Mol Sci. 2019. PMID: 30669312 Free PMC article. Review.
Cited by
- O-GlcNAc signaling attenuates ER stress-induced cardiomyocyte death.
Ngoh GA, Hamid T, Prabhu SD, Jones SP. Ngoh GA, et al. Am J Physiol Heart Circ Physiol. 2009 Nov;297(5):H1711-9. doi: 10.1152/ajpheart.00553.2009. Epub 2009 Sep 4. Am J Physiol Heart Circ Physiol. 2009. PMID: 19734355 Free PMC article. - Metabolic Stress and Cardiovascular Disease in Diabetes Mellitus: The Role of Protein _O_-GlcNAc Modification.
Chen Y, Zhao X, Wu H. Chen Y, et al. Arterioscler Thromb Vasc Biol. 2019 Oct;39(10):1911-1924. doi: 10.1161/ATVBAHA.119.312192. Epub 2019 Aug 29. Arterioscler Thromb Vasc Biol. 2019. PMID: 31462094 Free PMC article. Review. - The role of protein O-linked beta-N-acetylglucosamine in mediating cardiac stress responses.
Chatham JC, Marchase RB. Chatham JC, et al. Biochim Biophys Acta. 2010 Feb;1800(2):57-66. doi: 10.1016/j.bbagen.2009.07.004. Epub 2009 Jul 14. Biochim Biophys Acta. 2010. PMID: 19607882 Free PMC article. Review. - Mitochondrial _O_-GlcNAc Transferase Interacts with and Modifies Many Proteins and Its Up-Regulation Affects Mitochondrial Function and Cellular Energy Homeostasis.
Jóźwiak P, Ciesielski P, Zakrzewski PK, Kozal K, Oracz J, Budryn G, Żyżelewicz D, Flament S, Vercoutter-Edouart AS, Bray F, Lefebvre T, Krześlak A. Jóźwiak P, et al. Cancers (Basel). 2021 Jun 12;13(12):2956. doi: 10.3390/cancers13122956. Cancers (Basel). 2021. PMID: 34204801 Free PMC article.
References
- Torres CR, Hart GW. Topography and polypeptide distribution of terminal N-acetylglucosamine residues on the surfaces of intact lymphocytes. Evidence for O-linked GlcNAc. J Biol Chem. 1984;259:3308–3317. - PubMed
- Nandi A, Sprung R, Barma DK, Zhao Y, Kim SC, Falck JR, Zhao Y. Global identification of O-GlcNAc-modified proteins. Analytical chemistry. 2006;78:452–458. - PubMed
- Miller MW, Caracciolo MR, Berlin WK, Hanover JA. Phosphorylation and glycosylation of nucleoporins. Arch Biochem Biophys. 1999;367:51–60. - PubMed
- Slawson C, Housley MP, Hart GW. O-GlcNAc cycling: how a single sugar post-translational modification is changing the way we think about signaling networks. J Cell Biochem. 2006;97:71–83. - PubMed
- Zachara NE, O'Donnell N, Cheung WD, Mercer JJ, Marth JD, Hart GW. Dynamic O-GlcNAc modification of nucleocytoplasmic proteins in response to stress. A survival response of mammalian cells. J Biol Chem. 2004;279:30133–30142. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 083320/PHS HHS/United States
- R01 HL083320-04/HL/NHLBI NIH HHS/United States
- R01 HL094419/HL/NHLBI NIH HHS/United States
- R01 HL083320-03/HL/NHLBI NIH HHS/United States
- R01 HL083320-01A1/HL/NHLBI NIH HHS/United States
- R01 HL083320/HL/NHLBI NIH HHS/United States
- R01 HL083320-02/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases