EZH1 mediates methylation on histone H3 lysine 27 and complements EZH2 in maintaining stem cell identity and executing pluripotency - PubMed (original) (raw)
EZH1 mediates methylation on histone H3 lysine 27 and complements EZH2 in maintaining stem cell identity and executing pluripotency
Xiaohua Shen et al. Mol Cell. 2008.
Abstract
Trimethylation on H3K27 (H3K27me3) mediated by Polycomb repressive complex 2 (PRC2) has been linked to embryonic stem cell (ESC) identity and pluripotency. EZH2, the catalytic subunit of PRC2, has been reported as the sole histone methyltransferase that methylates H3K27 and mediates transcriptional silencing. Analysis of Ezh2(-/-) ESCs suggests existence of an additional enzyme(s) catalyzing H3K27 methylation. We have identified EZH1, a homolog of EZH2 that is physically present in a noncanonical PRC2 complex, as an H3K27 methyltransferase in vivo and in vitro. EZH1 colocalizes with the H3K27me3 mark on chromatin and preferentially preserves this mark on development-related genes in Ezh2(-/-) ESCs. Depletion of Ezh1 in cells lacking Ezh2 abolishes residual methylation on H3K27 and derepresses H3K27me3 target genes, demonstrating a role of EZH1 in safeguarding ESC identity. Ezh1 partially complements Ezh2 in executing pluripotency during ESC differentiation, suggesting that cell-fate transitions require epigenetic specificity.
Figures
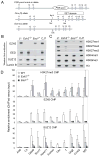
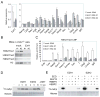
Figure 1. Altered methylation on H3K27 in _Ezh2_−/− cells
(A) Targeting strategy to generate Ezh2 conditional and knockout alleles. Exon number is indicated above each exon shown in boxes. Lines represent introns. (B–C) Western blot analysis of nuclear extracts (B) and histones (C) isolated from wild-type (J1 and CJ7) and mutant (_Ezh2_−/− and _Eed_−/−) ES cells. (D) ChIP-qPCR analysis of H3K27me3, EZH2 and SUZ12 binding at promoters of PcG target genes in wild-type (WT), _Ezh2_−/− and _Eed_−/− ES cells. The y-axis shows relative enrichment of ChIP’ed DNA versus the inputs. Actb promoter and an intergenetic region (Cs) serve as negative controls.
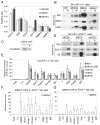
Figure 2. EZH1 interacts and co-localizes with EZH2 and EED at PcG target loci
(A) Proteins interacting with bfEZH2, bfEED or bfEZH1 were pulled down by FLAG and biotin-streptavidin (SA) mediated tandem coIP and subsequently identified by Mass Spec (MS) sequencing. Y-axis shows relative ratio of peptides representing an identified protein (x–axis) in total number of peptides identified by MS. Error bars represent standard deviations of ratios of peptides identified by MS in independent pull-down experiments. Neither bfEZH2 nor bfEZH1 pulls down each other (indicated by asterisks). (B) One-step SA coIP and Western blotting of J1 ES cells expressing birA alone or with any of bfEZH2, bfEZH1 and bfEED. “I” represents inputs and “P” represents coIP’ed fractions. (C) bfEZH1 is coIP’ed with endogenous EZH2. (D) SA coIP and Western blot analysis of _Ezh2_−/− ES cells expressing bfEZH1, bfEED or bfEZH2. (E) Colocalization of bfEZH1 with bfEZH2 and bfEED at promoters of PcG targets shown by bioChIP-qPCR analysis in J1 ES cells. (F–G) The binding of bfEZH1 (F) and bfEED (G) to PcG target loci in _Ezh2_−/− ES cells.
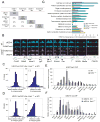
Figure 3. bfEZH1 colocalizes with H3K27me3 on chromatin
(A) The overlapping percentages of target genes identified by H3K27me3 and bfEZH1 ChIP-chip in wild-type (WT) and _Ezh2_−/− mutant (Mut) ES cells. The total numbers of target genes (p < 1e-7) is highlighted in grey. (B) Representative view of bfEZH1 and H3K27me3 occupancy at various loci in WT (shown in blue) and _Ezh2_−/− ES cells (in purple) by Affymetrix Integrated Genome Browser. Arrows indicate transcription starts and directions. Yellow bars indicate regions retaining strong binding in _Ezh2_−/− cells. Asterisks in red indicate regions losing ChIP peaks in _Ezh2_−/− cells. qPCR primers used in Panel E and F were designed in the above marked regions. (C–D) Comparisons of peak positions and widths of H3K27me3 (C) or bfEZH1 (D) ChIP-chip in _Ezh2_−/− and wild-type ES cells. Panel I shows deviations of mid-points of corresponding ChIP peaks in _Ezh2_−/− and wild-type cells. The distribution is centered at 0 and the deviations, by large part, are within 1 kb on both sides. Panel II shows the distribution of width differences of corresponding ChIP peaks in _Ezh2_−/− and wild-type cells. The distribution is heavily skewed to the left, indicating less broad peaks in _Ezh2_−/− cells than in wild-type cells in general. (E–F) qPCR verification of H3K27me3 ChIP (E) in WT and _Ezh2_−/− ES cells without exogenous bfEzh1 and bfEZH1 bioChIP (F) in cells expressing bfEzh1. (G) Gene Ontology (GO) analysis of H3K27me3 target genes. GO terms with an enrichment score larger than 2 (i.e. one-fold above genome background) and a _p_-value less than 1e-6 are considered significantly enriched and indicated by asterisks. The WTH3K27me3 gene set comprises of all H3K27me3 target genes. _Ezh2_−/−, H3K27me3+ or _Ezh2_−/−, H3K27me3− genes represent a subset of H3K27me3 target genes which retain or lose this mark in _Ezh2_−/− cells, respectively.
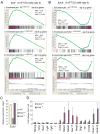
Figure 4. Ezh2 is dispensable for the maintenance of ES cell identity
(A–B) GSEA profiles of three subsets of H3K27me3 targets by comparing undifferentiated _Eed_−/−(A) or _Ezh2_−/− ES cells (B) to wild-type (WT) ES cells. Panel I, WTH3K27me3 – day 6 up genes; Panel II, _Ezh2_−/−, H3K27me3+ – day 6 up genes; Panel III, _Ezh2_−/−, H3K27me3− – day 6 up genes. All three subsets show positive normalized enrichment scores (NES) in _Eed_−/− ES cells but negative NES in _Ezh2_−/− ES cells, suggesting derepression in _Eed_−/− ES cells but repression in _Ezh2_−/− ES cells. (C) Ezh2 transcript is reduced ~ 1000 fold in _Ezh2_−/− cells compared to that in WT and _Eed_−/− cells. (D) Fold changes of gene expression in mutants versus wild-type cells.
Figure 5. Ezh1 mediates methylation on H3K27 in vivo and in vitro
(A–C) Ezh1 RNAi in _Ezh2_−/− ES cells. (A) RT-qPCR analysis of Ezh1 and marker genes. Relative expression was normalized to that in untreated _Ezh2_−/− ES cells. (B) Histone analysis. (C) Reduced H3K27me3 binding at PRC2 targets in Ezh1 RNAi cells shown by ChIP-qPCR. (D–E) HMTase assays. (D) FLAG-tagged EZH1, EZH2 or EZH2ΔSET was co-expressed with EED, SUZ12, RBBP4 and AEBP2 in Sf21 cells. (E) FLAG-tagged EZH1 or EZH2 was co-expressed with indicated PRC2 components in Sf21 cells. Protein complexes were pulled down by anti-FLAG agarose and assayed for HMTase activity. The upper panels show the autoradiography and the lower panels show equal addition of core histone substrates.
Figure 6. Ezh2 and Eed are required for mesoendodermal (ME) differentiation
(A) Heat map showing impaired activation of known mesodermal markers in _Ezh2_−/− and _Eed_−/−cells as compared to wild-type (WT) cells. (B) RT-qPCR analysis of T expression along ES cell differentiation by LIF withdrawal. (C–D) GSEA profiles of ME-high (panel I) and ES-high (panel II) genes by comparing day-6 differentiated _Eed_−/− (C) or _Ezh2_−/− (D) cells to wild-type cells. (E) Heat map of ME-high genes in wild-type and mutant ES cells. (F) RT-qPCR analysis of pluripotency markers along ES cell differentiation.
Figure 7. Complementary but non-redundant roles of EZH1 and EZH2 in mediating methylation on H3K27 and ES cell function
(A) A model in wild-type (WT) ES cells. Both EZH2 and EZH1 bind to EED and SUZ12, forming alternative but interacting PRC2 complexes containing HMTase activity. EZH2-mediated PRC2 is more abundant in ES cells and plays a major role in forming H3K27me3, which is required for ES cell identity and proper differentiation. Both EZH1 and EZH2 containing PRC2 complexes may mediate monomethylation on H3K27 in a non-targeted manner by either acting transiently on nucleosomes across the genome or on free H3 histones prior to nucleosomal assembly during DNA replication. (B) In _Ezh2_−/− ES cells, EZH1-PRC2 mediates monomethylation and residual trimethylation on H3K27. EED may regulate EZH1-PRC2 complex in an EZH2-dependant manner. EZH1 mediates H3K27me3 on a set of developmental genes and prevents derepression of PRC2 target genes from globe loss of the H3K27me3 mark in _Ezh2_−/− ES cells. Hereby, EZH1 complements EZH2 in the maintenance of ES cell identity. However, EZH1-PRC2 cannot fully rescue defects in mesoendodermal development during _Ezh2_−/− cell differentiation, suggesting non-redundant functions of these two PRC2 complexes in lineage commitment. (C) _Eed_−/− ES cells lack all levels of methylation on H3K27, indicating the essential role of EED in EZH1 or EZH2 mediated PRC2 complexes. Complete loss of H3K27me3 in the absence of Eed confounds ES cell identity by de-repressing a set of differentiation-regulated PRC2 target genes, and blocks the proper execution of pluripotent programs.
Similar articles
- Distinct Stimulatory Mechanisms Regulate the Catalytic Activity of Polycomb Repressive Complex 2.
Lee CH, Holder M, Grau D, Saldaña-Meyer R, Yu JR, Ganai RA, Zhang J, Wang M, LeRoy G, Dobenecker MW, Reinberg D, Armache KJ. Lee CH, et al. Mol Cell. 2018 May 3;70(3):435-448.e5. doi: 10.1016/j.molcel.2018.03.019. Epub 2018 Apr 19. Mol Cell. 2018. PMID: 29681498 Free PMC article. - EZH1 in germ cells safeguards the function of PRC2 during spermatogenesis.
Mu W, Starmer J, Shibata Y, Yee D, Magnuson T. Mu W, et al. Dev Biol. 2017 Apr 15;424(2):198-207. doi: 10.1016/j.ydbio.2017.02.017. Epub 2017 Feb 28. Dev Biol. 2017. PMID: 28254491 Free PMC article. - EZH1 and EZH2 cogovern histone H3K27 trimethylation and are essential for hair follicle homeostasis and wound repair.
Ezhkova E, Lien WH, Stokes N, Pasolli HA, Silva JM, Fuchs E. Ezhkova E, et al. Genes Dev. 2011 Mar 1;25(5):485-98. doi: 10.1101/gad.2019811. Epub 2011 Feb 11. Genes Dev. 2011. PMID: 21317239 Free PMC article. - The role of EZH1 and EZH2 in development and cancer.
Lee SH, Li Y, Kim H, Eum S, Park K, Lee CH. Lee SH, et al. BMB Rep. 2022 Dec;55(12):595-601. doi: 10.5483/BMBRep.2022.55.12.174. BMB Rep. 2022. PMID: 36476271 Free PMC article. Review. - EZH2 methyltransferase and H3K27 methylation in breast cancer.
Yoo KH, Hennighausen L. Yoo KH, et al. Int J Biol Sci. 2012;8(1):59-65. doi: 10.7150/ijbs.8.59. Epub 2011 Nov 18. Int J Biol Sci. 2012. PMID: 22211105 Free PMC article. Review.
Cited by
- An orally bioavailable chemical probe of the Lysine Methyltransferases EZH2 and EZH1.
Konze KD, Ma A, Li F, Barsyte-Lovejoy D, Parton T, Macnevin CJ, Liu F, Gao C, Huang XP, Kuznetsova E, Rougie M, Jiang A, Pattenden SG, Norris JL, James LI, Roth BL, Brown PJ, Frye SV, Arrowsmith CH, Hahn KM, Wang GG, Vedadi M, Jin J. Konze KD, et al. ACS Chem Biol. 2013;8(6):1324-34. doi: 10.1021/cb400133j. Epub 2013 Apr 24. ACS Chem Biol. 2013. PMID: 23614352 Free PMC article. - The Role of KDM2B and EZH2 in Regulating the Stemness in Colorectal Cancer Through the PI3K/AKT Pathway.
Sanches JGP, Song B, Zhang Q, Cui X, Yabasin IB, Ntim M, Li X, He J, Zhang Y, Mao J, Lu Y, Li L. Sanches JGP, et al. Front Oncol. 2021 Mar 9;11:637298. doi: 10.3389/fonc.2021.637298. eCollection 2021. Front Oncol. 2021. PMID: 33791221 Free PMC article. - Evaluation of histone 3 lysine 27 trimethylation (H3K27me3) and enhancer of Zest 2 (EZH2) in pediatric glial and glioneuronal tumors shows decreased H3K27me3 in H3F3A K27M mutant glioblastomas.
Venneti S, Garimella MT, Sullivan LM, Martinez D, Huse JT, Heguy A, Santi M, Thompson CB, Judkins AR. Venneti S, et al. Brain Pathol. 2013 Sep;23(5):558-64. doi: 10.1111/bpa.12042. Epub 2013 Mar 6. Brain Pathol. 2013. PMID: 23414300 Free PMC article. - _EZH2_-mediated H3K27me3 is a predictive biomarker and therapeutic target in uveal melanoma.
Hou C, Xiao L, Ren X, Cheng L, Guo B, Zhang M, Yan N. Hou C, et al. Front Genet. 2022 Oct 6;13:1013475. doi: 10.3389/fgene.2022.1013475. eCollection 2022. Front Genet. 2022. PMID: 36276954 Free PMC article. - Epigenetic regulation of the intestinal epithelium.
Elliott EN, Kaestner KH. Elliott EN, et al. Cell Mol Life Sci. 2015 Nov;72(21):4139-56. doi: 10.1007/s00018-015-1997-9. Epub 2015 Jul 29. Cell Mol Life Sci. 2015. PMID: 26220502 Free PMC article. Review.
References
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. - PubMed
- Bender LB, Cao R, Zhang Y, Strome S. The MES-2/MES-3/MES-6 complex and regulation of histone H3 methylation in C. elegans. Curr Biol. 2004;14:1639–1643. - PubMed
- Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials