Human UBN1 is an ortholog of yeast Hpc2p and has an essential role in the HIRA/ASF1a chromatin-remodeling pathway in senescent cells - PubMed (original) (raw)
Human UBN1 is an ortholog of yeast Hpc2p and has an essential role in the HIRA/ASF1a chromatin-remodeling pathway in senescent cells
Gowrishankar Banumathy et al. Mol Cell Biol. 2009 Feb.
Abstract
Cellular senescence is an irreversible proliferation arrest, tumor suppression process and likely contributor to tissue aging. Senescence is often characterized by domains of facultative heterochromatin, called senescence-associated heterochromatin foci (SAHF), which repress expression of proliferation-promoting genes. Given its likely contribution to tumor suppression and tissue aging, it is essential to identify all components of the SAHF assembly pathway. Formation of SAHF in human cells is driven by a complex of histone chaperones, namely, HIRA and ASF1a. In yeast, the complex orthologous to HIRA/ASF1a contains two additional proteins, Hpc2p and Hir3p. Using a sophisticated approach to search for remote orthologs conserved in multiple species through evolution, we identified the HIRA-associated proteins, UBN1 and UBN2, as candidate human orthologs of Hpc2p. We show that the Hpc2-related domain of UBN1, UBN2, and Hpc2p is an evolutionarily conserved HIRA/Hir-binding domain, which directly interacts with the N-terminal WD repeats of HIRA/Hir. UBN1 binds to proliferation-promoting genes that are repressed by SAHF and associates with histone methyltransferase activity that methylates lysine 9 of histone H3, a site that is methylated in SAHF. UBN1 is indispensable for formation of SAHF. We conclude that UBN1 is an ortholog of yeast Hpc2p and a novel regulator of senescence.
Figures
FIG. 1.
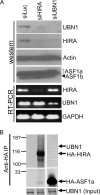
Identification of UBN1 and UBN2 as candidate Hpc2 orthologs. (A) Sequence alignment of S. cerevisiae Hpc2p and candidate orthologs from other species. Residues that are identical to Hpc2p are red, and those that are similar are yellow. At any position, at least 70% of residues must be similar for yellow to be applied. (B) Schematic full-length alignment of S. cerevisiae Hpc2p and human UBN1 and UBN2. The HRD from panel A is in pink. Other domains conserved between UBN1 and UBN2 are in yellow and blue. (C) UBN1 and UBN2 mRNAs are expressed in a panel of primary and transformed cell lines, based on RT-PCR analysis. Lane 1, primary WI38 fibroblasts; lane 2, primary IMR90 fibroblasts; lane 3, primary breast epithelial cells; lane 4, primary ovarian epithelial cells; lane 5, A498 renal carcinoma cells; lane 6, M14 melanoma cells; lane 7, MCF7 breast carcinoma cells; lane 8, OVCAR4, ovarian carcinoma cells; lane 9, HL60, leukemia cells. (D) Antibodies to UBN1 detect the UBN1 protein. Lanes 1 and 2, immunoprecipitation of cell extracts with control antibody or rabbit polyclonal pAbUBN1C; lanes 3 and 4, extracts from cells treated with the indicated siRNAs. All lanes were Western blotted with a mouse monoclonal antibody to UBN1. (E) Antibodies to ASF1a and UBN1 both (co)immunoprecipitate ASF1a, HIRA, and UBN1 from IMR90 fibroblasts. (F) Antibodies to UBN1 and HIRA both (co)immunoprecipitate UBN1, HIRA, and ASF1a from WI38 fibroblasts, hMPCs, and U2OS osteosarcoma cells. Input is 1/10 the amount of lysate used for immunoprecipitation. The bracket marks ASF1a, and the arrowhead marks the antibody light chain from the immunoprecipitation.
FIG. 2.
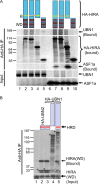
UBN1 interacts preferentially with HIRA. (A) IMR90 cells were nucleofected with siRNAs to luciferase, HIRA, or UBN1 as indicated. Cell extracts were Western blotted to detect UBN1, HIRA, and ASF1 (a mixture of anti-ASF1 polyclonal antibodies [pAb87 and -88] was used to detect both isoforms of ASF1, ASF1a and ASF1b). RNA was analyzed by RT-PCR for expression of HIRA, UBN1, and GAPDH (glyceraldehyde-3-phosphate dehydrogenase). (B) In vitro-translated 35S-labeled HA-tagged full-length HIRA or ASF1a was incubated with in vitro-translated 35S-labeled UBN1 and immunoprecipitated with anti-HA antibodies.
FIG. 3.
Mapping of HIRA and UBN1 interaction domains. (A) In vitro-translated 35S-labeled wild-type HA-tagged HIRA (lanes 2 and 7) or the indicated deletion mutants (lanes 3 to 5 and 8 to 10) were incubated with in vitro-translated 35S-labeled full-length wild-type UBN1 or ASF1a and immunoprecipitated with anti-HA antibodies. (B) In vitro-translated 35S-labeled wild-type HA-tagged UBN1 (lane 5) or the indicated UBN1 or UBN2 mutants (lanes 2 to 4) were incubated with in vitro-translated 35S-labeled HIRA(WD) (residues 1 to 400) and immunoprecipitated with anti-HA antibodies. The asterisks mark the HA-UBN1 or HA-UBN2 fusion proteins.
FIG. 4.
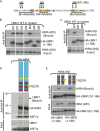
The HRD is an evolutionarily conserved HIRA-binding domain. (A) Sf9 cells were singly or coinfected with baculoviruses to express recombinant His-HIRA(1-405) and/or His-UBN1(1-175). His-tagged proteins were recovered from clarified extracts by IMAC and resolved on a Superdex 200 10/30 size exclusion column. The indicated globular molecular masses (670, 158, 44, and 17 kDa) provide an indication of the apparent hydrodynamic size of the complex but not a measurement of actual molecular mass. The void elution volume is indicated. (B) Purified recombinant GST or GST-Hir1p(WD) (residues 1 to 400) was incubated with 35S-labeled wild-type (Hpc2p) or mutant (Hpc2pΔHRD) Hpc2p as indicated. GST proteins and bound Hpc2p were isolated on glutathione beads.
FIG. 5.
Point mutations within the UBN1 HRD and the HIRA WD domain disrupt the HIRA/UBN1 and HIRA/UBN2 complexes. (A) The substitution mutants M1 to M8 that were tested for binding in panels B and C. Residues 124 to 170 of UBN1 are shown. (B) In vitro-translated 35S-labeled wild-type HA-tagged UBN1(1-166) or the mutants M1 to M6 were incubated with in vitro-translated 35S-labeled HIRA(WD) and immunoprecipitated with anti-HA antibodies. (C) As for panel B but with mutants M7 and M8 in HA-UBN1(1-166). (D) In vitro-translated 35S-labeled full-length wild-type HA-tagged human HIRA or HA-HIRA(R227K) was incubated with in vitro-translated 35S-labeled full-length UBN1 and ASF1a and then immunoprecipitated with anti-HA antibodies. (E) In vitro-translated 35S-labeled wild-type HA-tagged UBN1 or UBN2 was incubated with in vitro-translated 35S-labeled wild-type human HIRA(WD) or HIRA(WD, R227K) and immunoprecipitated with anti-HA antibodies.
FIG. 6.
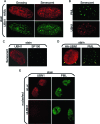
UBN1 is localized to PML bodies in senescent cells, together with HIRA. (A) Growing and senescent (after extended growth in culture) IMR90 fibroblasts were stained with antibodies to HIRA and UBN1. (B) Senescent (after extended growth in culture) IMR90 fibroblasts were stained with antibodies to PML and UBN1. (C) Senescent (after extended growth in culture) IMR90 fibroblasts were stained with antibodies to UBN1 and SP100. (D) IMR90 cells were infected with a retrovirus encoding HA-tagged UBN1 and stained with antibodies to HA and PML. Yellow arrows mark HA-UBN1 in PML bodies. (E) IMR90 cells were nucleofected with an siRNA to UBN1 or a control siRNA (siLuc) and then stained to detect UBN1 and PML.
FIG. 7.
UBN1 binds to the cyclin A2 gene and HMTase activity. (A) ChIP analysis of the cyclin A2 gene in proliferating IMR90 cells, with control (purified rabbit anti-mouse), anti-acetylated histone H3, or affinity-purified UBN1 antibodies as indicated. (B) Proliferating IMR90 cells were infected with a control retrovirus or a virus encoding HA-tagged UBN1 prior to ChIP analysis of the cyclin A2 gene using anti-HA antibodies. (C) Proliferating IMR90 cells were infected with a control lentivirus or a lentivirus encoding an shRNA to UBN1 and then subjected to ChIP analysis of the cyclin A2 gene with antibodies to UBN1 or control antibodies (melon gel purified from rabbit preimmune serum [PI]). (D) Proliferating IMR90 cells were nucleofected with control siRNA or siRNA to UBN1 and then subjected to ChIP analysis of the cyclin A2 gene with antibodies to UBN1 or control antibodies (PI). (E) ChIP analysis of the cyclin A2 gene, with control and anti-ASF1a antibodies as indicated. (F) Extracts were prepared from IMR90 cells and immunoprecipitated with control or anti-UBN1 antibodies. Immunoprecipitates were incubated with 3H-labeled _S_-adenosylmethionine and wild-type GST-histone H3 or the indicated GST-histone H3 mutants. The left two lanes were incubated with or without purified recombinant SETDB1 methyltransferase, as indicated. Top panel, autoradiograph; bottom panel, Coomassie blue stain.
FIG. 8.
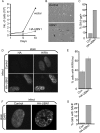
Ectopic expression of UBN1 induces senescence. (A) IMR90 cells were infected with a control retrovirus or a virus encoding HA-tagged wild-type UBN1. Infected cells were selected in puromycin. (B) Cells from panel A were stained for expression of senescence-associated (SA) β-galactosidase at 10 days after infection. (C) Quantitation of results from panel B. (D) Cells from panel A were stained with antibodies to HA and HIRA. (E) Quantitation of cells with HIRA foci from panel D. Error bars indicate standard deviations. (F) Cells from panel A were stained with DAPI. (G) Quantitation of cells with SAHF (DAPI foci) from panel F.
FIG. 9.
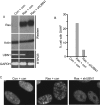
UBN1 is required for formation of SAHF. (A) IMR90 cells were infected with control retroviruses or viruses encoding activated Ras or shUBN1, as indicated, and then selected in puromycin. (B) Cells from panel A were scored for formation of SAHF. (C) Representative images of cells from panel A.
Similar articles
- Human CABIN1 is a functional member of the human HIRA/UBN1/ASF1a histone H3.3 chaperone complex.
Rai TS, Puri A, McBryan T, Hoffman J, Tang Y, Pchelintsev NA, van Tuyn J, Marmorstein R, Schultz DC, Adams PD. Rai TS, et al. Mol Cell Biol. 2011 Oct;31(19):4107-18. doi: 10.1128/MCB.05546-11. Epub 2011 Aug 1. Mol Cell Biol. 2011. PMID: 21807893 Free PMC article. - Identification of an ubinuclein 1 region required for stability and function of the human HIRA/UBN1/CABIN1/ASF1a histone H3.3 chaperone complex.
Tang Y, Puri A, Ricketts MD, Rai TS, Hoffmann J, Hoi E, Adams PD, Schultz DC, Marmorstein R. Tang Y, et al. Biochemistry. 2012 Mar 27;51(12):2366-77. doi: 10.1021/bi300050b. Epub 2012 Mar 16. Biochemistry. 2012. PMID: 22401310 Free PMC article. - Definition of pRB- and p53-dependent and -independent steps in HIRA/ASF1a-mediated formation of senescence-associated heterochromatin foci.
Ye X, Zerlanko B, Zhang R, Somaiah N, Lipinski M, Salomoni P, Adams PD. Ye X, et al. Mol Cell Biol. 2007 Apr;27(7):2452-65. doi: 10.1128/MCB.01592-06. Epub 2007 Jan 22. Mol Cell Biol. 2007. PMID: 17242198 Free PMC article. - Remodeling of chromatin structure in senescent cells and its potential impact on tumor suppression and aging.
Adams PD. Adams PD. Gene. 2007 Aug 1;397(1-2):84-93. doi: 10.1016/j.gene.2007.04.020. Epub 2007 May 1. Gene. 2007. PMID: 17544228 Free PMC article. Review. - A Molecular Prospective for HIRA Complex Assembly and H3.3-Specific Histone Chaperone Function.
Ricketts MD, Marmorstein R. Ricketts MD, et al. J Mol Biol. 2017 Jun 30;429(13):1924-1933. doi: 10.1016/j.jmb.2016.11.010. Epub 2016 Nov 19. J Mol Biol. 2017. PMID: 27871933 Free PMC article. Review.
Cited by
- HIRA, a conserved histone chaperone, plays an essential role in low-dose stress response via transcriptional stimulation in fission yeast.
Chujo M, Tarumoto Y, Miyatake K, Nishida E, Ishikawa F. Chujo M, et al. J Biol Chem. 2012 Jul 6;287(28):23440-50. doi: 10.1074/jbc.M112.349944. Epub 2012 May 15. J Biol Chem. 2012. PMID: 22589550 Free PMC article. - MEF2B Instructs Germinal Center Development and Acts as an Oncogene in B Cell Lymphomagenesis.
Brescia P, Schneider C, Holmes AB, Shen Q, Hussein S, Pasqualucci L, Basso K, Dalla-Favera R. Brescia P, et al. Cancer Cell. 2018 Sep 10;34(3):453-465.e9. doi: 10.1016/j.ccell.2018.08.006. Cancer Cell. 2018. PMID: 30205047 Free PMC article. - Whole Exome Sequencing of Highly Aggregated Lung Cancer Families Reveals Linked Loci for Increased Cancer Risk on Chromosomes 12q, 7p, and 4q.
Musolf AM, Moiz BA, Sun H, Pikielny CW, Bossé Y, Mandal D, de Andrade M, Gaba C, Yang P, Li Y, You M, Govindan R, Wilson RK, Kupert EY, Anderson MW, Schwartz AG, Pinney SM, Amos CI, Bailey-Wilson JE. Musolf AM, et al. Cancer Epidemiol Biomarkers Prev. 2020 Feb;29(2):434-442. doi: 10.1158/1055-9965.EPI-19-0887. Epub 2019 Dec 11. Cancer Epidemiol Biomarkers Prev. 2020. PMID: 31826912 Free PMC article. - HIRA complex presets transcriptional potential through coordinating depositions of the histone variants H3.3 and H2A.Z on the poised genes in mESCs.
Yang Y, Zhang L, Xiong C, Chen J, Wang L, Wen Z, Yu J, Chen P, Xu Y, Jin J, Cai Y, Li G. Yang Y, et al. Nucleic Acids Res. 2022 Jan 11;50(1):191-206. doi: 10.1093/nar/gkab1221. Nucleic Acids Res. 2022. PMID: 34893908 Free PMC article. - Structure of the Hir histone chaperone complex.
Kim HJ, Szurgot MR, van Eeuwen T, Ricketts MD, Basnet P, Zhang AL, Vogt A, Sharmin S, Kaplan CD, Garcia BA, Marmorstein R, Murakami K. Kim HJ, et al. Mol Cell. 2024 Jul 25;84(14):2601-2617.e12. doi: 10.1016/j.molcel.2024.05.031. Epub 2024 Jun 25. Mol Cell. 2024. PMID: 38925115
References
- Ait-Ahmed, O., B. Bellon, M. Capri, C. Joblet, and M. Thomas-Delaage. 1992. The yemanuclein-alpha: a new Drosophila DNA binding protein specific for the oocyte nucleus. Mech. Dev. 3769-80. - PubMed
- Berger, S. L. 2007. The complex language of chromatin regulation during transcription. Nature 447407-412. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM062281/GM/NIGMS NIH HHS/United States
- P01AG031862/AG/NIA NIH HHS/United States
- R01 CA129334-01/CA/NCI NIH HHS/United States
- P01 AG031862/AG/NIA NIH HHS/United States
- R01 CA129334/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases