The CTCF insulator protein is posttranslationally modified by SUMO - PubMed (original) (raw)
The CTCF insulator protein is posttranslationally modified by SUMO
Melissa J MacPherson et al. Mol Cell Biol. 2009 Feb.
Abstract
The CTCF protein is a highly conserved zinc finger protein that is implicated in many aspects of gene regulation and nuclear organization. Its functions include the ability to act as a repressor of genes, including the c-myc oncogene. In this paper, we show that the CTCF protein can be posttranslationally modified by the small ubiquitin-like protein SUMO. CTCF is SUMOylated both in vivo and in vitro, and we identify two major sites of SUMOylation in the protein. The posttranslational modification of CTCF by the SUMO proteins does not affect its ability to bind to DNA in vitro. SUMOylation of CTCF contributes to the repressive function of CTCF on the c-myc P2 promoter. We also found that CTCF and the repressive Polycomb protein, Pc2, are colocalized to nuclear Polycomb bodies. The Pc2 protein may act as a SUMO E3 ligase for CTCF, strongly enhancing its modification by SUMO 2 and 3. These studies expand the repertoire of posttranslational modifications of CTCF and suggest roles for such modifications in its regulation of epigenetic states.
Figures
FIG. 1.
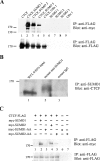
CTCF is SUMOylated by SUMO 1, 2, and 3, and SUMOylation depends on the SUMO protein's terminal diglycine residues. (A) CTCF is SUMOylated by SUMO 1, 2, and 3. HEK 293 cells were transfected with plasmids encoding FLAG-tagged CTCF and one of the three SUMO proteins or the ubiquitin-like protein Ufm1, all tagged with the myc epitope, as indicated above the top blot. (Top) The whole-cell lysates were immunoprecipitated (IP) with an antibody directed against the FLAG epitope (for CTCF), analyzed by SDS-PAGE and Western blotting, and probed with anti-myc antibody to detect SUMO or Ufm1 modifications. CTCF (>170,000-molecular-weight band) is modified by SUMO 1, 2, and 3 (lanes 2, 3, and 4) but not by Ufm1 (lane 5). Single transfections with CTCF and SUMO 1, 2, or 3 plasmids gave no signal. (Bottom) The same blot was stripped and reprobed with anti-FLAG antibody to show that CTCF was efficiently immunoprecipitated (lanes 1 to 5), whereas no signal was detected after transfection by SUMO- or Ufm1-encoding plasmids alone (lanes 6 to 9). (B) Endogenous CTCF is SUMOylated by SUMO 1. HEK 293 cells were harvested, lysed, and immunoprecipitated with antibody against SUMO 1 or mouse IgG as a control. The immunoprecipitates were analyzed by SDS-PAGE and Western blotting. Probing with anti-CTCF antibody detected a high-molecular-weight form of CTCF modified by SUMO 1 (lane 2). Whole-cell lysate (WCL) (0.85% of the input into each immunoprecipitation) was run in lane 1 to show the unmodified form of CTCF, which migrates faster through the SDS-PAGE gel than the SUMOylated form of CTCF (lane 2). (C) SUMOylation of CTCF by SUMO 1 and 2 is dependent on the two C-terminal glycine residues. HEK 293 cells were transfected with the plasmids indicated at the top. (Top) Whole-cell lysates were immunoprecipitated with anti-FLAG antibody, and Western blots were probed with anti-myc antibody. SUMOylated CTCF was detected after transfection with wild-type SUMO plasmids (lanes 2 and 4) but was absent or greatly reduced with the GG→AA mutant constructs (lanes 3 and 5). The bands were absent after transfection with single plasmids (lanes 1, 6, and 7). (Bottom) Probing the same blot with anti-FLAG antibody (for CTCF) showed that CTCF was efficiently immunoprecipitated.
FIG. 2.
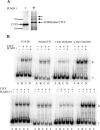
SUMOylation of CTCF is sensitive to the SENP1 and SENP5 isopeptidases. HEK 293 cells were transfected with plasmids encoding FLAG-tagged CTCF, one of the three _myc_-tagged SUMO proteins (lanes 2 to 10), and SENP1 (lanes 3, 6, and 9) or SENP5 (lanes 4, 7, and 10). (Top) Whole-cell lysates were immunoprecipitated (IP) with an anti-FLAG antibody (for CTCF), and the immunoprecipitates were subjected to SDS-PAGE, followed by Western blotting with an anti-myc antibody to detect the SUMO modifications. The SUMO 1 modification of CTCF (lane 2) was sensitive to SENP1 (lane 3) but not SENP5 (lane 4), whereas the modifications of CTCF by SUMO 2 and SUMO 3 (lanes 5 and 8) were sensitive to both SENP 1 (lanes 6 and 9) and SENP5 (lanes 7 and 10). This was the expected specificity of the SENPs. (Bottom) The same blot was reprobed with anti-FLAG antibody to show that CTCF was efficiently immunoprecipitated and that the proteases were not simply degrading CTCF nonspecifically.
FIG. 3.
SUMOylation of CTCF does not affect its DNA binding ability. (A) In-vitro translated 35S-labeled CTCF was incubated in a SUMOylation reaction without (−) and with (+) SUMO 1 protein and run on an SDS-PAGE gel. In the presence of SUMO 1, all of the CTCF was SUMOylated and shifted into more slowly migrating species (the arrows show the modified and unmodified forms of CTCF). Similar experiments also showed efficient SUMOylation by SUMO 2 and 3 (not shown). (B) CTCF was made by in vitro transcription/translation (IVT) and used in electrophoretic mobility shift assays with or without subsequent incubation in an in vitro SUMOylation reaction. The PCR fragments used in each experiment are shown and are described in Materials and Methods and Table 2. Lanes A, IVT mixture without CTCF mRNA; lanes B, IVT using CTCF mRNA; lanes C, IVT CTCF incubated in SUMOylation reaction mixture but without SUMO protein; lanes D, E, and F, IVT CTCF incubated in SUMOylation reaction mixture with SUMO 1, 2, or 3, respectively. U, unbound DNA; B, bound DNA. Careful inspection of lanes containing the SUMO proteins revealed a slight slowing of the shifted band, presumably due to SUMOylation of CTCF. Competition experiments showed that SUMOylation might affect more weakly binding sites (data not shown).
FIG. 4.
Identification of SUMOylation sites in CTCF. (A) The amino acid sequence of mouse CTCF protein is shown with the SUMOylation sites highlighted in the N terminus and the C terminus of the protein. The boldface serine residues were previously described by others as being posttranslationally modified by phosphorylation. (B) The C-terminal and N-terminal domains of CTCF are SUMOylated in vitro. GST-tagged N- and C-terminal domains of CTCF were expressed in E. coli, purified, and incubated in a SUMOylation reaction with (+) and without (−) SUMO 1 protein. The reactions were run on an SDS-PAGE gel, subjected to Western blotting, and probed with antibodies to the N terminus (term) or C terminus of CTCF. Purified SUMO 1 protein (lanes 2 and 4) effectively SUMOylated both domains. (C) CTCF is SUMOylated at the V
K
KE and M
K
TE motifs. HEK 293 cells were transfected with wild-type (WT) FLAG-CTCF or single-mutant FLAG-CTCF V
R
KE or double-mutant V
R
KE M
R
TE plasmids (as indicated above the gel), in addition to plasmids encoding _myc_-tagged SUMO 1, 2, or 3 (as indicated below the gel). After immunoprecipitation (IP) with anti-FLAG antibodies, SDS-PAGE, and Western blotting, the membranes were probed with anti-myc antibodies for SUMO (top) or with anti-FLAG antibodies for CTCF (bottom). The V
R
KE 698 mutation markedly diminished SUMOylation by SUMO 1, 2, and 3 (lanes 3, 6, and 9). The double mutations M
R
TE 74 and V
R
KE 698 diminished the residual signal (lanes 4, 7, and 10).
FIG. 5.
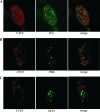
Colocalization of CTCF and Pc2 proteins in HeLa cell nuclei. HeLa cells were fixed in paraformaldehyde and stained with antibodies. (A) Cells were stained with mouse antibody to CTCF (red) and rabbit antibody to Pc2 protein (green) and counterstained with anti-mouse Texas Red-labeled antibody and anti-rabbit fluorescein isothiocyanate-labeled antibody. The merged image shows clear colocalization of CTCF and Pc2 (yellow). The Pearson coefficient for the colocalization was calculated using ImageJ and showed a high level of colocalization at 0.829. A value of 1 or −1 is considered perfect colocalization. (B) Cells were stained with goat antibody to CTCF (red) and mouse antibody to PML (green) and counterstained with anti-goat Alexa 488-labeled antibody and anti-mouse Alexa 647-labeled antibody. The merged image shows that CTCF and PML did not clearly colocalize. The Pearson coefficient for the colocalization was 0.375. These results cannot be interpreted as partial colocalization or as exclusion, since midrange coefficients between 0.5 and −0.5 are inconclusive (7). (C) Staining with goat antibody to CTCF (red) and mouse antibody to SC-35 (green) with counterstaining as in panel B. The merged image shows that CTCF and SC-35 do not colocalize and are found in different subnuclear compartments. The Pearson coefficient for the colocalization was −0.008, which is close to zero, an indication of exclusion. The images were generated using confocal laser scanning microscopy. Our protocol used nonionic detergent to permeabilize the fixed cells, and it is possible that the detergent treatment of the cells might have removed a soluble fraction of CTCF from the nucleus.
FIG. 6.
The SUMO E3 ligase Pc2 increases the SUMOylation of CTCF. HeLa cells were transfected using Fugene 6 transfection reagent with the indicated plasmids. The whole-cell lysates were precleared with anti-T7-agarose resin prior to immunoprecipitation (IP) with anti-FLAG antibodies. After SDS-PAGE and Western blotting, the membrane was probed with anti-myc antibody to detect SUMO 1, 2, or 3 (top) or with anti-FLAG antibody to detect CTCF (middle). SUMO E3 ligase activity was indicated by the increase in the SUMOylation signal in lanes 3 and 11. Pc2 exhibited no SUMO E3 ligase activity for the SUMOylation of CTCF by SUMO 1 (lane 6). CTCF degradation products were also SUMOylated by SUMO 2 and 3 in the presence of Pc2. Flowthrough from the immunoprecipitation was blotted for β-actin to show equal loading (bottom).
FIG. 7.
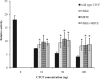
SUMOylation of CTCF contributes to its repressive effect on the c-myc P2 promoter. HEK 293 cells were transfected with 500 ng of the c-myc P2 luciferase reporter construct either with no added CTCF-encoding plasmid (far left black bar) or with 10, 50, or 100 ng of plasmid DNAs encoding either wild-type CTCF or CTCF bearing mutations in the SUMO modification sites. The luciferase activities were measured as described in Materials and Methods and were normalized to the activity of a cotransfected plasmid encoding Renilla luciferase. The repressive effect of CTCF on the promoter is shown by the concentration-dependent four- to fivefold reduction in luciferase activity (black bars). This reduction was partially decreased by either the single V
R
KE or M
R
TE mutation or the double V
R
KE and M
R
TE mutations, which decrease the SUMOylation of CTCF. The asterisks indicate that the results were statistically significant (P < 0.05) when a pairwise Student t test was used to compare the CTCF mutants to the wild-type conditions.
Similar articles
- The polycomb protein Pc2 is a SUMO E3.
Kagey MH, Melhuish TA, Wotton D. Kagey MH, et al. Cell. 2003 Apr 4;113(1):127-37. doi: 10.1016/s0092-8674(03)00159-4. Cell. 2003. PMID: 12679040 - c-Myc is targeted to the proteasome for degradation in a SUMOylation-dependent manner, regulated by PIAS1, SENP7 and RNF4.
González-Prieto R, Cuijpers SA, Kumar R, Hendriks IA, Vertegaal AC. González-Prieto R, et al. Cell Cycle. 2015;14(12):1859-72. doi: 10.1080/15384101.2015.1040965. Cell Cycle. 2015. PMID: 25895136 Free PMC article. - MEL-18 interacts with HSF2 and the SUMO E2 UBC9 to inhibit HSF2 sumoylation.
Zhang J, Goodson ML, Hong Y, Sarge KD. Zhang J, et al. J Biol Chem. 2008 Mar 21;283(12):7464-9. doi: 10.1074/jbc.M707122200. Epub 2008 Jan 21. J Biol Chem. 2008. PMID: 18211895 Free PMC article. - Pc2 and SUMOylation.
Wotton D, Merrill JC. Wotton D, et al. Biochem Soc Trans. 2007 Dec;35(Pt 6):1401-4. doi: 10.1042/BST0351401. Biochem Soc Trans. 2007. PMID: 18031231 Review. - [Identification and function of ubiquitin-like protein SUMO E3 (PIAS family and RanBp2, Pc2)].
Takahashi Y. Takahashi Y. Seikagaku. 2004 Apr;76(4):381-4. Seikagaku. 2004. PMID: 15162967 Review. Japanese. No abstract available.
Cited by
- Genome-wide studies of CCCTC-binding factor (CTCF) and cohesin provide insight into chromatin structure and regulation.
Lee BK, Iyer VR. Lee BK, et al. J Biol Chem. 2012 Sep 7;287(37):30906-13. doi: 10.1074/jbc.R111.324962. Epub 2012 Sep 5. J Biol Chem. 2012. PMID: 22952237 Free PMC article. Review. - Enhancer-promoter interactions are encoded by complex genomic signatures on looping chromatin.
Whalen S, Truty RM, Pollard KS. Whalen S, et al. Nat Genet. 2016 May;48(5):488-96. doi: 10.1038/ng.3539. Epub 2016 Apr 4. Nat Genet. 2016. PMID: 27064255 Free PMC article. - Mechanisms of TP53 Pathway Inactivation in Embryonic and Somatic Cells-Relevance for Understanding (Germ Cell) Tumorigenesis.
Timmerman DM, Remmers TL, Hillenius S, Looijenga LHJ. Timmerman DM, et al. Int J Mol Sci. 2021 May 20;22(10):5377. doi: 10.3390/ijms22105377. Int J Mol Sci. 2021. PMID: 34065345 Free PMC article. Review. - The SUMO protease SENP1 and the chromatin remodeler CHD3 interact and jointly affect chromatin accessibility and gene expression.
Rodríguez-Castañeda F, Lemma RB, Cuervo I, Bengtsen M, Moen LM, Ledsaak M, Eskeland R, Gabrielsen OS. Rodríguez-Castañeda F, et al. J Biol Chem. 2018 Oct 5;293(40):15439-15454. doi: 10.1074/jbc.RA118.002844. Epub 2018 Aug 6. J Biol Chem. 2018. PMID: 30082317 Free PMC article. - Epigenetic silencing of the XAF1 gene is mediated by the loss of CTCF binding.
Victoria-Acosta G, Vazquez-Santillan K, Jimenez-Hernandez L, Muñoz-Galindo L, Maldonado V, Martinez-Ruiz GU, Melendez-Zajgla J. Victoria-Acosta G, et al. Sci Rep. 2015 Oct 7;5:14838. doi: 10.1038/srep14838. Sci Rep. 2015. PMID: 26443201 Free PMC article.
References
- Anckar, J., and L. Sistonen. 2007. SUMO: getting it on. Biochem. Soc. Trans. 351409-1413. - PubMed
- Andersson, S., D. L. Davis, H. Dahlback, H. Jornvall, and D. W. Russell. 1989. Cloning, structure, and expression of the mitochondrial cytochrome P-450 sterol 26-hydroxylase, a bile acid biosynthetic enzyme. J. Biol. Chem. 2648222-8229. - PubMed
- Barski, A., S. Cuddapah, K. Cui, T. Y. Roh, D. E. Schones, Z. Wang, G. Wei, I. Chepelev, and K. Zhao. 2007. High-resolution profiling of histone methylations in the human genome. Cell 129823-837. - PubMed
- Bell, A. C., and G. Felsenfeld. 2000. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature 405482-485. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases