Analysis of combinatorial cis-regulation in synthetic and genomic promoters - PubMed (original) (raw)
Analysis of combinatorial cis-regulation in synthetic and genomic promoters
Jason Gertz et al. Nature. 2009.
Abstract
Transcription factor binding sites are being discovered at a rapid pace. It is now necessary to turn attention towards understanding how these sites work in combination to influence gene expression. Quantitative models that accurately predict gene expression from promoter sequence will be a crucial part of solving this problem. Here we present such a model, based on the analysis of synthetic promoter libraries in yeast (Saccharomyces cerevisiae). Thermodynamic models based only on the equilibrium binding of transcription factors to DNA and to each other captured a large fraction of the variation in expression in every library. Thermodynamic analysis of these libraries uncovered several phenomena in our system, including cooperativity and the effects of weak binding sites. When applied to the S. cerevisiae genome, a model of repression by Mig1 (which was trained on synthetic promoters) predicts a number of Mig1-regulated genes that lack significant Mig1-binding sites in their promoters. The success of the thermodynamic approach suggests that the information encoded by combinations of cis-regulatory sites is interpreted primarily through simple protein-DNA and protein-protein interactions, with complicated biochemical reactions-such as nucleosome modifications-being downstream events. Quantitative analyses of synthetic promoter libraries will be an important tool in unravelling the rules underlying combinatorial cis-regulation.
Figures
Figure 1
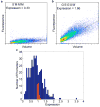
Gene expression measurements. Graphs of cell volume versus fluorescence for 25,000 individual cells containing the promoters A) S′MM′M and B) G′S′G′S′M′ where S = Spacer, G = Gcr1 site, M = Mig1 site and the ′ superscript indicates a site in the reverse orientation. C) Histogram of expression values for all L1 library members. Expression values were computed as the average fluorescence/volume ratio for 25,000 individual cells, and then normalized to plate controls. Control promoters with no library insert are shown in red.
Figure 2
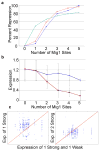
Mig1 sites act cooperatively and weak site represses weakly. A) Hill equation with _n_=3.4 and _K_=1.8 (red) fits the observed data (blue) well compared to _n_=1 (green). B) Plot of average expression versus the number of weak sites (blue), without strong sites, and strong sites (red), without weak sites. Error bars represent one standard deviation. C) Plots of expression for pairs of identical promoters except that either one strong Mig1 site or two strong Mig1 sites replace one strong and one weak Mig1 site. A blue circle represents one promoter pair and the red line represents equal expression.
Figure 3
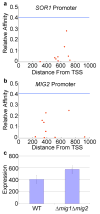
Thermodynamic model explains Mig1 repression in the genome. Mig1 binding sites in the promoters of SOR1 (A) and MIG2 (B) are shown. The affinity of Mig1 for the site based on a position weight matrix score relative to the strong site is plotted vs. the location upstream on the translation start site (TSS). The horizontal line represents the significance threshold for the weight matrix and each square represents a Mig1 site. C) MIG2 promoter activity in a wild-type (WT) strain and a mig1Δ mig2Δ strain, error bars represent one standard deviation.
Similar articles
- TATA is a modular component of synthetic promoters.
Mogno I, Vallania F, Mitra RD, Cohen BA. Mogno I, et al. Genome Res. 2010 Oct;20(10):1391-7. doi: 10.1101/gr.106732.110. Epub 2010 Jul 13. Genome Res. 2010. PMID: 20627890 Free PMC article. - Combinatorial control of gene expression by the three yeast repressors Mig1, Mig2 and Mig3.
Westholm JO, Nordberg N, Murén E, Ameur A, Komorowski J, Ronne H. Westholm JO, et al. BMC Genomics. 2008 Dec 16;9:601. doi: 10.1186/1471-2164-9-601. BMC Genomics. 2008. PMID: 19087243 Free PMC article. - A yeast transcription assay defines distinct rel and dorsal DNA recognition sequences.
Kamens J, Brent R. Kamens J, et al. New Biol. 1991 Oct;3(10):1005-13. New Biol. 1991. PMID: 1768648 - Genome-wide analysis of the context-dependence of regulatory networks.
Papp B, Oliver S. Papp B, et al. Genome Biol. 2005;6(2):206. doi: 10.1186/gb-2005-6-2-206. Epub 2005 Jan 27. Genome Biol. 2005. PMID: 15693953 Free PMC article. Review. - Synthetic Promoters: Designing the cis Regulatory Modules for Controlled Gene Expression.
Aysha J, Noman M, Wang F, Liu W, Zhou Y, Li H, Li X. Aysha J, et al. Mol Biotechnol. 2018 Aug;60(8):608-620. doi: 10.1007/s12033-018-0089-0. Mol Biotechnol. 2018. PMID: 29855997 Review.
Cited by
- Mechanistic analysis of enhancer sequences in the estrogen receptor transcriptional program.
Tabe-Bordbar S, Song YJ, Lunt BJ, Alavi Z, Prasanth KV, Sinha S. Tabe-Bordbar S, et al. Commun Biol. 2024 Jun 11;7(1):719. doi: 10.1038/s42003-024-06400-5. Commun Biol. 2024. PMID: 38862711 Free PMC article. - Regulatory properties of transcription factors with diverse mechanistic function.
Ali MZ, Guharajan S, Parisutham V, Brewster RC. Ali MZ, et al. PLoS Comput Biol. 2024 Jun 10;20(6):e1012194. doi: 10.1371/journal.pcbi.1012194. eCollection 2024 Jun. PLoS Comput Biol. 2024. PMID: 38857275 Free PMC article. - Proformer: a hybrid macaron transformer model predicts expression values from promoter sequences.
Kwak IY, Kim BC, Lee J, Kang T, Garry DJ, Zhang J, Gong W. Kwak IY, et al. BMC Bioinformatics. 2024 Feb 20;25(1):81. doi: 10.1186/s12859-024-05645-5. BMC Bioinformatics. 2024. PMID: 38378442 Free PMC article. - Transcription factor interactions explain the context-dependent activity of CRX binding sites.
Loell KJ, Friedman RZ, Myers CA, Corbo JC, Cohen BA, White MA. Loell KJ, et al. PLoS Comput Biol. 2024 Jan 16;20(1):e1011802. doi: 10.1371/journal.pcbi.1011802. eCollection 2024 Jan. PLoS Comput Biol. 2024. PMID: 38227575 Free PMC article. - Hold out the genome: a roadmap to solving the cis-regulatory code.
de Boer CG, Taipale J. de Boer CG, et al. Nature. 2024 Jan;625(7993):41-50. doi: 10.1038/s41586-023-06661-w. Epub 2023 Dec 13. Nature. 2024. PMID: 38093018 Review.
References
- Hu Z, Killion PJ, Iyer VR. Genetic reconstruction of a functional transcriptional regulatory network. Nature genetics. 2007;39:683–687. - PubMed
- Beer MA, Tavazoie S. Predicting gene expression from sequence. Cell. 2004;117:185–198. - PubMed
- Bussemaker HJ, Li H, Siggia ED. Regulatory element detection using correlation with expression. Nature genetics. 2001;27:167–171. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases