Nutrient sensing and inflammation in metabolic diseases - PubMed (original) (raw)
Review
Nutrient sensing and inflammation in metabolic diseases
Gökhan S Hotamisligil et al. Nat Rev Immunol. 2008 Dec.
Abstract
The proper functioning of the pathways that are involved in the sensing and management of nutrients is central to metabolic homeostasis and is therefore among the most fundamental requirements for survival. Metabolic systems are integrated with pathogen-sensing and immune responses, and these pathways are evolutionarily conserved. This close functional and molecular integration of the immune and metabolic systems is emerging as a crucial homeostatic mechanism, the dysfunction of which underlies many chronic metabolic diseases, including type 2 diabetes and atherosclerosis. In this Review we provide an overview of several important networks that sense and manage nutrients and discuss how they integrate with immune and inflammatory pathways to influence the physiological and pathological metabolic states in the body.
Conflict of interest statement
Competing interests statement
The author(s) declare(s) competing financial interests: see web version for details.
Figures
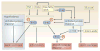
Figure 1. molecular characteristics shared between adipocytes and macrophages in physiological conditions and metabolic disease states
An extensive transcriptional signature is common to adipocytes and macrophages. The Venn diagram shows the number of genes that are expressed preferentially or approximately equally by pre-adipocytes, adipocytes and macrophages. Inflammatory genes that are expressed by both macrophages and adipocytes are also listed, as are adipocyte-specific metabolic and other genes that are upregulated during the transformation of macrophages into foam cells–,,.
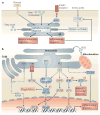
Figure 2. nutrient sensing and inflammation
The signalling pathways that sense and respond to three basic nutrients, free fatty acids, glucose and amino acids, are depicted. Many pathways that monitor nutrient availability, such as those of mammalian target of rapamycin (mTOR), insulin and nuclear transcription factors (not shown), and the unfolded-protein response (UPR) in the endoplasmic reticulum (ER), closely interact with each other. They also interact with inflammatory pathways, particularly those that are induced through the activation of IκB kinase-β (IKKβ) and JUN N-terminal kinase (JNK). Leptin resistance can be induced through ER stress and through the activation of the IKK pathway when overnutrition is detected by the hypothalamus,. In addition to these intracellular points of crosstalk, nutrients may directly induce inflammation through pathogen-sensing receptors, such as the activation of Toll-like receptor (TLR) signalling by free fatty acids. Dysregulation of the crosstalk between the pathways that are shown here can culminate in altered metabolic responses, inflammation and leptin resistance. IκB, inhibitor of nuclear factor-κB; IRS1, insulin receptor substrate 1; FABP, fatty-acid binding protein; TNF, tumour-necrosis factor.
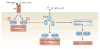
Figure 3. The unfolded-protein response, nutrient sensing and inflammation
a. The endoplasmic reticulum (ER) is an important organelle that responds to multiple nutrient-associated signals, such as those induced by fatty acids, glucose, free cholesterol, insulin and amino acids. The unfolded-protein response (UPR) is induced in response to ER stress and activates stress-response pathways (not shown) and inflammatory signalling pathways that can result in altered metabolic and inflammatory responses and consequently in metabolic disease. The inflammatory response can enhance the UPR and further activate mammalian target of rapamycin (mTOR). b. The UPR is mediated by three different pathways that are initiated by three transmembrane proteins that are located in the ER — activating transcription factor 6 (ATF6), pancreatic ER kinase (PERK) and inositol-requiring kinase 1 (IRE1). ER stress is linked to inflammation through the activation of the JUN N-terminal kinase (JNK) and the IκB kinase (IKK)–nuclear factor-κB (NFκB) pathways, and through cyclic-AMP-responsive-element-binding protein H (CREBH) activation by the UPR. These pathways result in the induction of an inflammatory response. Activation of JNK can also serine phosphorylate insulin receptor substrate 1 (IRS1), resulting in altered metabolic responses. Key organelles for cellular metabolism, such as the ER, Golgi, mitochondria and peroxisomes (not shown) are connected through an endomembrane network, which provides functional continuity between organelles that can therefore share functional information in the form of lipids and proteins at specific contact sites. This functional and molecular integration between the organelles can mediate the spread of stress from one organelle to the other, resulting in exacerbation of inflammation and cytotoxicity during chronic metabolic stress conditions such as obesity and dyslipidaemia. AP1, activator protein 1; ATF6f, ATF6 fragment; eIF2α, eukaryotic translation initiation factor 2α; IκB, inhibitor of NFκB; XBP1s, spliced X-box binding protein 1.
Figure 4. The mammalian target of rapamycin pathway, amino-acid sensing and inflammation
Mammalian target of rapamycin (mTOR) is an important checkpoint kinase that transmits signals related to amino-acid sufficiency and protein synthesis. A hyperactive mTOR pathway has been associated with increased signalling induced by the unfolded-protein response (UPR) and with the activation of JUN N-terminal kinase (JNK), which can lead to increased inflammation and insulin resistance through serine phosphorylation of insulin receptor substrate 1 (IRS1). Inflammation can further activate mTOR, through IκB kinase (IKK)-mediated phosphorylation of tuberous sclerosis complex 1 (TSC1), as can the UPR through the induction of activating transcription factor 6 (ATF6; not shown). This leads to a vicious inflammatory cycle in metabolically stressed cells. The UPR and IKK activation can also lead to leptin resistance through the induction of polypyrimidine tract-binding protein 1β (PTB1β) and suppressor of cytokine signalling 3 (SOCS3), respectively. 4EBP1, eukaryotic translation-initiation factor 4E-binding protein 1; GβL, G-protein β-subunit-like protein; IκB, inhibitor of nuclear factor-κB; mTORC2, mTOR complex 2; RAPTOR, regulatory associated protein of mTOR; RHEB, RAS homology enriched in brain; S6K1, ribosomal protein S6 kinase 1; TNF, tumour-necrosis factor.
Figure 5. lipid-sensing pathways and inflammation
Increased amounts of fatty acids can directly induce Toll-like receptors (TLRs) and lipid sensors or transmit stress signals following binding to intracellular lipid chaperones known as fatty-acid binding proteins (FABPs). Activation of TLRs induces the activation of IκB kinase (IKK) and JUN N-terminal kinase (JNK). JNK can also be activated as a result of endoplasmic reticulum (ER) stress inside the cell (not shown). The ER responds to increased levels of lipotoxic fatty acids and free cholesterol by activating the unfolded-protein response. Activation of two transcription factors that are associated with the ER, X-box binding protein 1 (XBP1s) and sterol-regulatory-element-binding protein (SREBP), can modify intracellular lipid metabolism. Furthermore, fatty acids can directly serve as both activating and inhibiting ligands for nuclear receptors, such as peroxisome-proliferator-activated receptors (PPARs) and liver X receptors (LXRs), and affect inflammation.
Similar articles
- Foundations of Immunometabolism and Implications for Metabolic Health and Disease.
Hotamisligil GS. Hotamisligil GS. Immunity. 2017 Sep 19;47(3):406-420. doi: 10.1016/j.immuni.2017.08.009. Immunity. 2017. PMID: 28930657 Free PMC article. Review. - Metabolic Flexibility as an Adaptation to Energy Resources and Requirements in Health and Disease.
Smith RL, Soeters MR, Wüst RCI, Houtkooper RH. Smith RL, et al. Endocr Rev. 2018 Aug 1;39(4):489-517. doi: 10.1210/er.2017-00211. Endocr Rev. 2018. PMID: 29697773 Free PMC article. Review. - Inflammation, metaflammation and immunometabolic disorders.
Hotamisligil GS. Hotamisligil GS. Nature. 2017 Feb 8;542(7640):177-185. doi: 10.1038/nature21363. Nature. 2017. PMID: 28179656 Review. - Inflammation and metabolic disorders.
Hotamisligil GS. Hotamisligil GS. Nature. 2006 Dec 14;444(7121):860-7. doi: 10.1038/nature05485. Nature. 2006. PMID: 17167474 Review. - Nutrient sensors and their crosstalk.
Sung Y, Yu YC, Han JM. Sung Y, et al. Exp Mol Med. 2023 Jun;55(6):1076-1089. doi: 10.1038/s12276-023-01006-z. Epub 2023 Jun 1. Exp Mol Med. 2023. PMID: 37258576 Free PMC article. Review.
Cited by
- Atf-6 Regulates Lifespan through ER-Mitochondrial Calcium Homeostasis.
Burkewitz K, Feng G, Dutta S, Kelley CA, Steinbaugh M, Cram EJ, Mair WB. Burkewitz K, et al. Cell Rep. 2020 Sep 8;32(10):108125. doi: 10.1016/j.celrep.2020.108125. Cell Rep. 2020. PMID: 32905769 Free PMC article. - Dietary Supplementation with Nucleotides, Short-Chain Fructooligosaccharides, Xylooligosaccharides, Beta-Carotene and Vitamin E Influences Immune Function in Kittens.
Atwal J, Joly W, Bednall R, Albanese F, Farquhar M, Holcombe LJ, Watson P, Harrison M. Atwal J, et al. Animals (Basel). 2023 Dec 2;13(23):3734. doi: 10.3390/ani13233734. Animals (Basel). 2023. PMID: 38067085 Free PMC article. - Chemokines in Prediabetes and Type 2 Diabetes: A Meta-Analysis.
Pan X, Kaminga AC, Wen SW, Liu A. Pan X, et al. Front Immunol. 2021 May 13;12:622438. doi: 10.3389/fimmu.2021.622438. eCollection 2021. Front Immunol. 2021. PMID: 34054797 Free PMC article. - FADD is a key regulator of lipid metabolism.
Zhuang H, Wang X, Zha D, Gan Z, Cai F, Du P, Yang Y, Yang B, Zhang X, Yao C, Zhou Y, Jiang C, Guan S, Zhang X, Zhang J, Jiang W, Hu Q, Hua ZC. Zhuang H, et al. EMBO Mol Med. 2016 Aug 1;8(8):895-918. doi: 10.15252/emmm.201505924. Print 2016 Aug. EMBO Mol Med. 2016. PMID: 27357657 Free PMC article. - Extrahepatic Malignancies in Nonalcoholic Fatty Liver Disease.
Ahmed OT, Allen AM. Ahmed OT, et al. Curr Hepatol Rep. 2019 Dec;18(4):455-472. doi: 10.1007/s11901-019-00499-5. Epub 2019 Nov 15. Curr Hepatol Rep. 2019. PMID: 36397965 Free PMC article.
References
- Hotamisligil GS. Inflammation and metabolic disorders. Nature. 2006;444:860–867. - PubMed
- Demas GE, Chefer V, Talan MI, Nelson RJ. Metabolic costs of mounting an antigen-stimulated immune response in adult and aged C57BL/56J mice. Am J Physiol. 1997;273:R1631–R1637. - PubMed
- Marti A, Marcos A, Martinez JA. Obesity and immune function relationships. Obes Rev. 2001;2:131–140. - PubMed
- Romanyukha AA, Rudnev SG, Sidorov IA. Energy cost of infection burden: an approach to understanding the dynamics of host-pathogen interactions. J Theor Biol. 2006;241:1–13. - PubMed
- Browning RC, Kram R. Energetic cost and preferred speed of walking in obese vs. normal weight women. Obes Res. 2005;13:891–899. - PubMed
Publication types
MeSH terms
Grants and funding
- DK64360/DK/NIDDK NIH HHS/United States
- R01 DK052539-11/DK/NIDDK NIH HHS/United States
- R01 HL065405-05/HL/NHLBI NIH HHS/United States
- R01 DK064360-05A1/DK/NIDDK NIH HHS/United States
- R01 DK056894-03/DK/NIDDK NIH HHS/United States
- R01 HL065405-07/HL/NHLBI NIH HHS/United States
- R01 DK054017-03/DK/NIDDK NIH HHS/United States
- R01 HL065405-02/HL/NHLBI NIH HHS/United States
- DK52539/DK/NIDDK NIH HHS/United States
- R01 DK052539-12/DK/NIDDK NIH HHS/United States
- R01 DK052539-02/DK/NIDDK NIH HHS/United States
- R01 HL065405-09/HL/NHLBI NIH HHS/United States
- R01 DK056894-04/DK/NIDDK NIH HHS/United States
- R01 DK052539-08/DK/NIDDK NIH HHS/United States
- R01 DK064360-02/DK/NIDDK NIH HHS/United States
- R01 HL065405-03/HL/NHLBI NIH HHS/United States
- R01 DK064360-05A1S1/DK/NIDDK NIH HHS/United States
- R01 DK064360/DK/NIDDK NIH HHS/United States
- R01 DK054017-02/DK/NIDDK NIH HHS/United States
- R01 DK052539-07/DK/NIDDK NIH HHS/United States
- HL65405/HL/NHLBI NIH HHS/United States
- R01 DK052539-10/DK/NIDDK NIH HHS/United States
- R01 DK052539-03/DK/NIDDK NIH HHS/United States
- R01 HL065405-06/HL/NHLBI NIH HHS/United States
- R01 HL065405-01/HL/NHLBI NIH HHS/United States
- R01 DK064360-01A2/DK/NIDDK NIH HHS/United States
- R01 HL065405-04/HL/NHLBI NIH HHS/United States
- R01 DK056894-01A1/DK/NIDDK NIH HHS/United States
- R01 HL065405/HL/NHLBI NIH HHS/United States
- R01 DK052539-06/DK/NIDDK NIH HHS/United States
- R01 DK052539/DK/NIDDK NIH HHS/United States
- R01 DK052539-04/DK/NIDDK NIH HHS/United States
- R56 DK064360/DK/NIDDK NIH HHS/United States
- R01 DK056894-02/DK/NIDDK NIH HHS/United States
- R01 DK064360-04/DK/NIDDK NIH HHS/United States
- R01 DK052539-09/DK/NIDDK NIH HHS/United States
- R01 HL065405-08/HL/NHLBI NIH HHS/United States
- R01 DK054017-04/DK/NIDDK NIH HHS/United States
- R01 DK052539-06S1/DK/NIDDK NIH HHS/United States
- R56 DK064360-05/DK/NIDDK NIH HHS/United States
- R01 DK056894/DK/NIDDK NIH HHS/United States
- R01 DK052539-05/DK/NIDDK NIH HHS/United States
- R01 DK064360-03/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical