Differential inhibitory activities and stabilisation of DNA aptamers against the SARS coronavirus helicase - PubMed (original) (raw)
Differential inhibitory activities and stabilisation of DNA aptamers against the SARS coronavirus helicase
Ka To Shum et al. Chembiochem. 2008.
Abstract
The helicase from severe acute respiratory syndrome coronavirus (SARS-CoV) possesses NTPase, duplex RNA/DNA-unwinding and RNA-capping activities that are essential for viral replication and proliferation. Here, we have isolated DNA aptamers against the SARS-CoV helicase from a combinatorial DNA library. These aptamers show two distinct classes of secondary structure, G-quadruplex and non-G-quadruplex, as shown by circular dichroism and gel electrophoresis. All of the aptamers that were selected stimulated ATPase activity of the SARS-CoV helicase with low-nanomolar apparent K(m) values. Intriguingly, only the non-G-quadruplex aptamers showed specific inhibition of helicase activities, whereas the G-quadruplex aptamers did not inhibit helicase activities. The non-G-quadruplex aptamer with the strongest inhibitory potency was modified at the 3'-end with biotin or inverted thymidine, and the modification increased its stability in serum, particularly for the inverted thymidine modification. Structural diversity in selection coupled to post-selection stabilisation has provided new insights into the aptamers that were selected for a helicase target. These aptamers are being further developed to inhibit SARS-CoV replication.
Figures
Figure 1
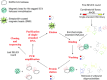
DNA aptamer selection strategy. His‐tagged SARS‐CoV helicase was immobilised on Ni‐NTA magnetic beads. The library was incubated with the target beads for binding. Unbound oligonucleotides were washed away, and the bound ones were eluted with the target by imidazole. The selected binders were amplified by PCR by using biotinylated primers. ssDNA was subsequently purified from the PCR product, resulting in an enriched DNA pool, which was used in the next SELEX round. After the last round, the selected aptamers were cloned, sequenced and characterised.
Figure 2
Sequences of the aptamers that were isolated from the ssDNA pool after twenty rounds of selection against SARS‐CoV helicase. Two groups of sequences were classified by the presence of G‐quadruplex structure that was predicted by QGRS mapper. A) Multiple sequence alignment of non‐G‐quadruplex aptamers by clustalW2. B) Multiple sequence alignment of G‐quadruplex‐forming aptamers. Guanine nucleotides that participated in formation of G‐quadruplex structure were predicted by QGRS mapper and are in bold type‐face and underlined.
Figure 3
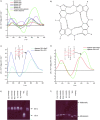
Secondary structure analysis of selected DNA aptamers. A) CD spectrometry was performed for the putative non‐G‐quadruplex aptamers NG1, NG3 and NG8 and putative G‐quadruplex aptamers G5 and G8 in 10 m
m
Tris–HCl buffer pH 7.5 and 100 m
m
KCl buffer solution. The concentration of oligonucleotides was 10 μ
m
. B) G‐quadruplex structure. Four guanine bases interact in a square planar configuration to form a G‐quadruplex. Each base interacts with adjacent bases through two hydrogen bonds by Hoogsteen‐like hydrogen bonding. CD spectra of C) aptamer G5 and D) aptamer G8 measured in the presence of either NaCl or KCl buffer solution. E) Mobility of the fluorescently labelled oligonucleotides in a 20 % polyacrylamide gel containing 8
m
urea. F) Mobility of folded aptamers on 16 % native polyacrylamide gel supplemented with 50 m
m
KCl.
Figure 4
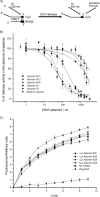
Inhibition of SARS‐CoV helicase partial duplex DNA unwinding activity. A) Schematic showing the principles behind the FRET‐based fluorimetric assay of helicase activity. B) Unwinding was performed in the presence of various concentrations of the five different aptamers and one random sequence. C) Observation of helicase activity of E. coli DnaB in the absence and presence of 1 μ
m
or 5 μ
m
aptamer NG1, NG3 and NG8. BSA (35 μg) was added as a negative control. The FRET assay was used to observe helicase activity by fluorescence over time. Points shown are an average of triplicate experiments. Error bars represents the standard deviation of triplicate measurements.
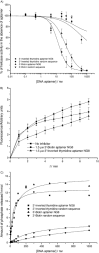
Figure 5
Determination of the apparent strength of binding of aptamers to the SARS‐CoV helicase by hydrolysis of ATP in the presence of varying concentrations of different aptamers. A colorimetric assay was used to measure the amount of phosphate release due to ATP to ADP hydrolysis. Data were shown after subtraction of basal ATPase phosphate release and the ATPase activities in the presence of each aptamer were fitted to a simple Michaelis–Menten model by using average values from three independent determinations.
Figure 6
Stability of 3′‐inverted thymidine‐, 3′‐biotin‐modified and unmodified NG8 in 5 and 10 % FBS. Aptamer NG8 with various modifications was mixed with 5 or 10 % FBS and incubated at 37 °C. Aliquots of the reaction mixture were taken at different time points and loaded into 20 % denaturing urea PAGE; A) 3′‐inverted thymidine aptamer NG8; B) 3′‐biotin aptamer NG8 and C) unmodified aptamer NG8 in 5 % FBS; D) 3′‐inverted thymidine aptamer NG8; E) 3′‐biotin aptamer NG8 and F) unmodified aptamer NG8 in 10 % FBS.
Figure 7
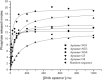
Biochemical assays of modified aptamers NG8. A) Inhibition of helicase activity of the SCV helicase in the presence of various concentrations of the modified aptamers. B) Specificity of modified aptamers by using E. coli DnaB. C) Effect of modified aptamers on the ATPase activity of SCV helicase.
Similar articles
- Severe acute respiratory syndrome coronavirus replication inhibitor that interferes with the nucleic acid unwinding of the viral helicase.
Adedeji AO, Singh K, Calcaterra NE, DeDiego ML, Enjuanes L, Weiss S, Sarafianos SG. Adedeji AO, et al. Antimicrob Agents Chemother. 2012 Sep;56(9):4718-28. doi: 10.1128/AAC.00957-12. Epub 2012 Jun 25. Antimicrob Agents Chemother. 2012. PMID: 22733076 Free PMC article. - Identification of myricetin and scutellarein as novel chemical inhibitors of the SARS coronavirus helicase, nsP13.
Yu MS, Lee J, Lee JM, Kim Y, Chin YW, Jee JG, Keum YS, Jeong YJ. Yu MS, et al. Bioorg Med Chem Lett. 2012 Jun 15;22(12):4049-54. doi: 10.1016/j.bmcl.2012.04.081. Epub 2012 Apr 25. Bioorg Med Chem Lett. 2012. PMID: 22578462 Free PMC article. - Isolation of inhibitory RNA aptamers against severe acute respiratory syndrome (SARS) coronavirus NTPase/Helicase.
Jang KJ, Lee NR, Yeo WS, Jeong YJ, Kim DE. Jang KJ, et al. Biochem Biophys Res Commun. 2008 Feb 15;366(3):738-44. doi: 10.1016/j.bbrc.2007.12.020. Epub 2007 Dec 17. Biochem Biophys Res Commun. 2008. PMID: 18082623 Free PMC article. - G-quadruplex DNA aptamers and their ligands: structure, function and application.
Tucker WO, Shum KT, Tanner JA. Tucker WO, et al. Curr Pharm Des. 2012;18(14):2014-26. doi: 10.2174/138161212799958477. Curr Pharm Des. 2012. PMID: 22376117 Review. - Development of chemical inhibitors of the SARS coronavirus: viral helicase as a potential target.
Keum YS, Jeong YJ. Keum YS, et al. Biochem Pharmacol. 2012 Nov 15;84(10):1351-8. doi: 10.1016/j.bcp.2012.08.012. Epub 2012 Aug 23. Biochem Pharmacol. 2012. PMID: 22935448 Free PMC article. Review.
Cited by
- Aptamer-decorated nanocarriers for viral adsorption: A special look at COVID-19.
Handali S, Rezaei M. Handali S, et al. Mol Ther Nucleic Acids. 2024 Aug 15;35(3):102310. doi: 10.1016/j.omtn.2024.102310. eCollection 2024 Sep 10. Mol Ther Nucleic Acids. 2024. PMID: 39281706 Free PMC article. Review. - Enhanced SELEX Platforms for Aptamer Selection with Improved Characteristics: A Review.
Didarian R, Ozbek HK, Ozalp VC, Erel O, Yildirim-Tirgil N. Didarian R, et al. Mol Biotechnol. 2024 Aug 16. doi: 10.1007/s12033-024-01256-w. Online ahead of print. Mol Biotechnol. 2024. PMID: 39152308 Review. - Coronaviruses SARS-CoV, MERS-CoV, and SARS-CoV-2 helicase inhibitors: a systematic review of in vitro studies.
Mehyar N. Mehyar N. J Virus Erad. 2023 Jun;9(2):100327. doi: 10.1016/j.jve.2023.100327. Epub 2023 May 26. J Virus Erad. 2023. PMID: 37363132 Free PMC article. Review. - Nanostructures for prevention, diagnosis, and treatment of viral respiratory infections: from influenza virus to SARS-CoV-2 variants.
Sharifi E, Yousefiasl S, Trovato M, Sartorius R, Esmaeili Y, Goodarzi H, Ghomi M, Bigham A, Moghaddam FD, Heidarifard M, Pourmotabed S, Nazarzadeh Zare E, Paiva-Santos AC, Rabiee N, Wang X, Tay FR. Sharifi E, et al. J Nanobiotechnology. 2023 Jun 21;21(1):199. doi: 10.1186/s12951-023-01938-8. J Nanobiotechnology. 2023. PMID: 37344894 Free PMC article. Review. - An overview on medicinal plants used for combating coronavirus: Current potentials and challenges.
Abou Baker DH, Hassan EM, El Gengaihi S. Abou Baker DH, et al. J Agric Food Res. 2023 Sep;13:100632. doi: 10.1016/j.jafr.2023.100632. Epub 2023 May 20. J Agric Food Res. 2023. PMID: 37251276 Free PMC article.
References
- Kuiken T., Fouchier R. A., Schutten M., Rimmelzwaan G. F., van Amerongen G., van Riel D., Laman J. D., de Jong T., van Doornum G., Lim W., Ling A. E., Chan P. K., Tam J. S., Zambon M. C., Gopal R., Drosten C., van der Werf S., Escriou N., Manuguerra J. C., Stohr K., Peiris J. S., Osterhaus A. D., Lancet 2003, 362, 263–270. - PMC - PubMed
- Ksiazek T. G., Erdman D., Goldsmith C. S., Zaki S. R., Peret T., Emery S., Tong S., Urbani C., Comer J. A., Lim W., Rollin P. E., Dowell S. F., Ling A. E., Humphrey C. D., Shieh W. J., Guarner J., Paddock C. D., Rota P., Fields B., DeRisi J., Yang J. Y., Cox N., Hughes J. M., LeDuc J. W., Bellini W. J., Anderson L. J., N. Engl. J. Med. 2003, 348, 1953–1966. - PubMed
- Rota P. A., Oberste M. S., Monroe S. S., Nix W. A., Campagnoli R., Icenogle J. P., Penaranda S., Bankamp B., Maher K., Chen M. H., Tong S., Tamin A., Lowe L., Frace M., DeRisi J. L., Chen Q., Wang D., Erdman D. D., Peret T. C., Burns C., Ksiazek T. G., Rollin P. E., Sanchez A., Liffick S., Holloway B., Limor J., McCaustland K., Olsen‐Rasmussen M., Fouchier R., Gunther S., Osterhaus A. D., Drosten C., Pallansch M. A., Anderson L. J., Bellini W. J., Science 2003, 300, 1394–1399. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous