A large genome center's improvements to the Illumina sequencing system - PubMed (original) (raw)
A large genome center's improvements to the Illumina sequencing system
Michael A Quail et al. Nat Methods. 2008 Dec.
Abstract
The Wellcome Trust Sanger Institute is one of the world's largest genome centers, and a substantial amount of our sequencing is performed with 'next-generation' massively parallel sequencing technologies: in June 2008 the quantity of purity-filtered sequence data generated by our Genome Analyzer (Illumina) platforms reached 1 terabase, and our average weekly Illumina production output is currently 64 gigabases. Here we describe a set of improvements we have made to the standard Illumina protocols to make the library preparation more reliable in a high-throughput environment, to reduce bias, tighten insert size distribution and reliably obtain high yields of data.
Figures
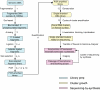
Figure 1. Illumina sequencing workflow
Stages in the library prep. Steps accompanied by numbers are those for which we suggest alternatives to the standard Illumina protocols and numbers correspond to those given in Supplementary Protocols 1-13 online.
Figure 2. Sample Fragmentation
Comparison of fragmentation by nebulization with AFA technology. 4.5μg of human genomic DNA was fragmented by nebulization and AFA, purified with a spin column, eluted in 30μl of 10mM Tris pH8.5 and 1μl of each eluate was run on an Agilent Bioanalyzer 2100 DNA 100 chip. For a 200bp library +/- 20bp, the yield produced by AFA is four- to fivefold greater than that produced by nebulization.
Figure 3. A-tailing, ligation and size selection
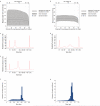
GC plots before (a) and after (b) optimisation of gel extraction. The figures show the total area in which reads with a particular GC content are distributed, with the mean and standard deviation. The greater width of shaded area in plot a) indicates a wider dispersion of coverage for all values of GC content for which sequences were obtained. Agilent traces Bioanalyzer 2100 traces for two suboptimal libraries c) 60bp insert library, with optimised PCR, d) the same 60bp library with excess DNA in PCR e) 200bp insert library, showing shoulder of small fragments. Insert size distribution from sequenced human DNA using f) the standard and g) modified paired end library prep protocols
Figure 4. PCR
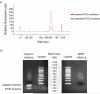
a) A ∼200bp fragment library was prepared, and 10ng was amplified for 18 cycles using standard Illumina conditions, and with more optimal PCR conditions. A comparison of methods of PCR amplicon purification. We prepared a paired end library with phiX DNA using conditions that would promote the formation of adapter dimers and unextended PCR primers. After PCR we divided the library into two: half was purified following the standard Illumina protocol, through a Qiaquick PCR cleanup column, whereas the other was purified using SPRI technology. Each was then run on an agarose gel alongside a 100bp ladder to view the DNA species that remained.
Figure 5. Quantification
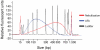
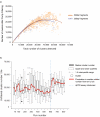
a) Cluster throughput as a function of total clusters for 200 and 500bp inserts. The 500bp inserts underwent fewer cycles of cluster amplification (28, compared to 35 for the 200bp libraries), resulting in smaller clusters, and so a cluster density of 40-44k / tile (GA1) will produce the maximum yield from either insert size. b) Standardisation of cluster density with qPCR quantification. Runs were grouped into 25-run bins and a boxplot plotted. After some initial problems with degradation of standards, cluster number has levelled out at ∼35-40k / tile.
Figure 6. Denaturation
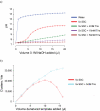
a) pH titration of hybridisation buffers. Following denaturation, the concentration of NaOH in DNA templates is 0.1M NaOH. Adding more than 8μl of this denatured template to the 1ml of Hybridisation Buffer (5x SSC, 0.1% Tween-20), prior to loading DNA onto the flowcell, increases the pH to above 10. This prevents efficient hybridisation, and thus the cluster density falls. The addition of Tris-HCl pH7.3 to the supplied bottles of Hybridisation Buffer dramatically increases buffering capacity, making template hybridisation more robust. b) the addition of 5mM Tris-HCl pH 7.3 to Illumina Hybridisation Buffer allows a greater volume of denatured template to be added before high pH prevents effective annealing of templates to the oligos on the flowcell surface. This increases the robustness of cluster generation, by counteracting pipetting errors in the denaturation step.
Similar articles
- The development and impact of 454 sequencing.
Rothberg JM, Leamon JH. Rothberg JM, et al. Nat Biotechnol. 2008 Oct;26(10):1117-24. doi: 10.1038/nbt1485. Nat Biotechnol. 2008. PMID: 18846085 - Improved protocols for the illumina genome analyzer sequencing system.
Quail MA, Swerdlow H, Turner DJ. Quail MA, et al. Curr Protoc Hum Genet. 2009 Jul;Chapter 18:Unit 18.2. doi: 10.1002/0471142905.hg1802s62. Curr Protoc Hum Genet. 2009. PMID: 19582764 Free PMC article. - Light-speed genomics.
Eisenstein M. Eisenstein M. Nat Methods. 2005 Sep;2(9):646. doi: 10.1038/nmeth0905-646. Nat Methods. 2005. PMID: 16184669 - Producing parasitic helminth reference and draft genomes at the Wellcome Trust Sanger Institute.
Holroyd N, Sanchez-Flores A. Holroyd N, et al. Parasite Immunol. 2012 Feb-Mar;34(2-3):100-7. doi: 10.1111/j.1365-3024.2011.01311.x. Parasite Immunol. 2012. PMID: 21707658 Review. - Next-generation DNA sequencing techniques.
Ansorge WJ. Ansorge WJ. N Biotechnol. 2009 Apr;25(4):195-203. doi: 10.1016/j.nbt.2008.12.009. Epub 2009 Feb 3. N Biotechnol. 2009. PMID: 19429539 Review.
Cited by
- Conserved DNA methylation patterns in healthy blood cells and extensive changes in leukemia measured by a new quantitative technique.
Jelinek J, Liang S, Lu Y, He R, Ramagli LS, Shpall EJ, Estecio MR, Issa JP. Jelinek J, et al. Epigenetics. 2012 Dec 1;7(12):1368-78. doi: 10.4161/epi.22552. Epub 2012 Oct 17. Epigenetics. 2012. PMID: 23075513 Free PMC article. - Elucidation of the RamA regulon in Klebsiella pneumoniae reveals a role in LPS regulation.
De Majumdar S, Yu J, Fookes M, McAteer SP, Llobet E, Finn S, Spence S, Monahan A, Monaghan A, Kissenpfennig A, Ingram RJ, Bengoechea J, Gally DL, Fanning S, Elborn JS, Schneiders T. De Majumdar S, et al. PLoS Pathog. 2015 Jan 29;11(1):e1004627. doi: 10.1371/journal.ppat.1004627. eCollection 2015 Jan. PLoS Pathog. 2015. PMID: 25633080 Free PMC article. - Ultrasensitive measurement of hotspot mutations in tumor DNA in blood using error-suppressed multiplexed deep sequencing.
Narayan A, Carriero NJ, Gettinger SN, Kluytenaar J, Kozak KR, Yock TI, Muscato NE, Ugarelli P, Decker RH, Patel AA. Narayan A, et al. Cancer Res. 2012 Jul 15;72(14):3492-8. doi: 10.1158/0008-5472.CAN-11-4037. Epub 2012 May 10. Cancer Res. 2012. PMID: 22581825 Free PMC article. - Adapter dimer contamination in sRNA-sequencing datasets predicts sequencing failure and batch effects and hampers extracellular vesicle-sRNA analysis.
Maqueda JJ, Giovanazzi A, Rocha AM, Rocha S, Silva I, Saraiva N, Bonito N, Carvalho J, Maia L, Wauben MHM, Oliveira C. Maqueda JJ, et al. J Extracell Biol. 2023 Jun 11;2(6):e91. doi: 10.1002/jex2.91. eCollection 2023 Jun. J Extracell Biol. 2023. PMID: 38938917 Free PMC article. - Genome-wide chromatin immunoprecipitation-sequencing in Plasmodium.
Lopez-Rubio JJ, Siegel TN, Scherf A. Lopez-Rubio JJ, et al. Methods Mol Biol. 2013;923:321-33. doi: 10.1007/978-1-62703-026-7_23. Methods Mol Biol. 2013. PMID: 22990789 Free PMC article.
References
- Bentley DR. Whole-genome re-sequencing. Curr Opin Genet Dev. 2006;16:545–552. - PubMed
- Mardis ER. The impact of next-generation sequencing technology on genetics. Trends Genet. 2008;24:133–141. - PubMed
- Surzycki S. Basic Techniques in Molecular Biology. Springer-Verlag; Berlin: 2000. pp. 377–380.
- Albert TJ, et al. Direct selection of human genomic loci by microarray hybridization. Nat Methods. 2007;4:903–905. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources