Phylogenetic evidence for extensive lateral acquisition of cellular genes by Nucleocytoplasmic large DNA viruses - PubMed (original) (raw)
Phylogenetic evidence for extensive lateral acquisition of cellular genes by Nucleocytoplasmic large DNA viruses
Jonathan Filée et al. BMC Evol Biol. 2008.
Abstract
Background: Nucleo-Cytoplasmic Large DNA viruses (NCLDV), a diverse group that infects a wide range of eukaryotic hosts, exhibit a large heterogeneity in genome size (between 100 kb and 1.2 Mb) but have been suggested to form a monophyletic group on the basis of a small subset of approximately 30 conserved genes. NCLDV were proposed to have evolved by simplification from cellular organism although some of the giant NCLDV have clearly grown by gene accretion from a bacterial origin.
Results: We demonstrate here that many NCLDV lineages appear to have undergone frequent gene exchange in two different ways. Viruses which infect protists directly (Mimivirus) or algae which exist as intracellular protists symbionts (Phycodnaviruses) acquire genes from a bacterial source. Metazoan viruses such as the Poxviruses show a predominant acquisition of host genes. In both cases, the laterally acquired genes show a strong tendency to be positioned at the tip of the genome. Surprisingly, several core genes believed to be ancestral in the family appear to have undergone lateral gene transfers, suggesting that the NCLDV ancestor might have had a smaller genome than previously believed. Moreover, our data show that the larger the genome, the higher is the number of laterally acquired genes. This pattern is incompatible with a genome reduction from a cellular ancestor.
Conclusion: We propose that the NCLDV viruses have evolved by significant growth of a simple DNA virus by gene acquisition from cellular sources.
Figures
Figure 1
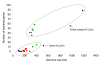
Number of bacterial-like genes in the NCLDVs. The number of genes identified as bacterial-like (excluding those of mobile elements) for each virus is plotted as a function of their genome size. Poxviruses are indicated by red circles, Iridoviruses are in blue, the Asfarvirus in orange, Phycodnaviruses in green and the Mimivirus in black.
Figure 2
Distribution of BLAST score for NCLDV ORFs with eukaryotic affinities. Upper panels: The axes represent the best BLASTP score against the host genome (horizontal) and against a non-redundant (NR) database purged from the closely related sequences of the host. The black lines correspond to equal scores along both axes. Lower panels: The axes represent the ratio: BLAST score against the host divided by the BLAST score against a NR database (vertical) plotted as the genome position of the corresponding ORF. The black lines correspond to the ratio equal to 1. A) Fowlpox BLAST score (best hit in the Bird sequence database against a NR database purged of vertebrate sequences). B) Amsacta moorei Poxvirus BLAST score (best hit in the Insect sequence database against a NR database purged of insect sequences). C) Lumpyskin disease Poxvirus (best hit in the Bos Taurus sequence database against a NR database purged of vertebrate sequences). D) Trichoplusia ni Ascovirus (Iridovirus) (best hit in Insect sequence database against a NR database purged of insect sequences). E) Frog Iridovirus (best hit in the Xenopus laevis sequence database against a NR database purged of vertebrate sequences). F) PBCV1 Chlorella phycodnavirus (best hit in the green alga and plant sequence database against a NR database purged of green alga and plant sequences).
Figure 3
Maximum likelihood phylogeny of several core NCLDV genes that display lateral gene transfers from the host. A) ATP-dependant DNA ligase. B) dUTPase. C) Serine/thréonine kinase. D) Thymidine kinase. E) Ribonucleotide reductase (small subunit). F) Ribonucleotide reductase (large subunit). Viral sequences are indicated in italics. Brackets represent putative cases of horizontal gene transfers between the virus and its host. Bootstrap values up to 95% are indicated with black circle. The scale bars represent the number of amino acid substitutions per residue.
Figure 4
Number of host derived genes in the NCLDVs. The number of host derived genes for each virus is plotted as a function of their genome size. Poxviruses are indicated with a red circle, Iridoviruses are in blue, the Asfarvirus in orange, Phycodnaviruses in green and the Mimivirus in black.
Similar articles
- Evolutionary genomics of nucleo-cytoplasmic large DNA viruses.
Iyer LM, Balaji S, Koonin EV, Aravind L. Iyer LM, et al. Virus Res. 2006 Apr;117(1):156-84. doi: 10.1016/j.virusres.2006.01.009. Epub 2006 Feb 21. Virus Res. 2006. PMID: 16494962 Review. - Origin and evolution of eukaryotic large nucleo-cytoplasmic DNA viruses.
Koonin EV, Yutin N. Koonin EV, et al. Intervirology. 2010;53(5):284-92. doi: 10.1159/000312913. Epub 2010 Jun 15. Intervirology. 2010. PMID: 20551680 Free PMC article. - Eukaryotic large nucleo-cytoplasmic DNA viruses: clusters of orthologous genes and reconstruction of viral genome evolution.
Yutin N, Wolf YI, Raoult D, Koonin EV. Yutin N, et al. Virol J. 2009 Dec 17;6:223. doi: 10.1186/1743-422X-6-223. Virol J. 2009. PMID: 20017929 Free PMC article. - Evolution of DNA ligases of nucleo-cytoplasmic large DNA viruses of eukaryotes: a case of hidden complexity.
Yutin N, Koonin EV. Yutin N, et al. Biol Direct. 2009 Dec 18;4:51. doi: 10.1186/1745-6150-4-51. Biol Direct. 2009. PMID: 20021668 Free PMC article. - Evolution of the Large Nucleocytoplasmic DNA Viruses of Eukaryotes and Convergent Origins of Viral Gigantism.
Koonin EV, Yutin N. Koonin EV, et al. Adv Virus Res. 2019;103:167-202. doi: 10.1016/bs.aivir.2018.09.002. Epub 2018 Nov 10. Adv Virus Res. 2019. PMID: 30635076 Review.
Cited by
- Structures of giant icosahedral eukaryotic dsDNA viruses.
Xiao C, Rossmann MG. Xiao C, et al. Curr Opin Virol. 2011 Aug;1(2):101-9. doi: 10.1016/j.coviro.2011.06.005. Curr Opin Virol. 2011. PMID: 21909343 Free PMC article. Review. - Evolution of viruses and cells: do we need a fourth domain of life to explain the origin of eukaryotes?
Moreira D, López-García P. Moreira D, et al. Philos Trans R Soc Lond B Biol Sci. 2015 Sep 26;370(1678):20140327. doi: 10.1098/rstb.2014.0327. Philos Trans R Soc Lond B Biol Sci. 2015. PMID: 26323758 Free PMC article. - Unraveling gene content variation across eukaryotic giant viruses based on network analyses and host associations.
Sun TW, Ku C. Sun TW, et al. Virus Evol. 2021 Sep 16;7(2):veab081. doi: 10.1093/ve/veab081. eCollection 2021. Virus Evol. 2021. PMID: 34754514 Free PMC article. - Cryopreservation of Paramecium bursaria Chlorella Virus-1 during an active infection cycle of its host.
Coy SR, Alsante AN, Van Etten JL, Wilhelm SW. Coy SR, et al. PLoS One. 2019 Mar 14;14(3):e0211755. doi: 10.1371/journal.pone.0211755. eCollection 2019. PLoS One. 2019. PMID: 30870463 Free PMC article. - Comparative Genomic Analysis of Acanthamoeba Endosymbionts Highlights the Role of Amoebae as a "Melting Pot" Shaping the Rickettsiales Evolution.
Wang Z, Wu M. Wang Z, et al. Genome Biol Evol. 2017 Nov 1;9(11):3214-3224. doi: 10.1093/gbe/evx246. Genome Biol Evol. 2017. PMID: 29177480 Free PMC article.
References
- Luria SE, Darnell JE. , JrGeneral virology. New York. 1967.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous