Gamma-secretase limits the inflammatory response through the processing of LRP1 - PubMed (original) (raw)
Gamma-secretase limits the inflammatory response through the processing of LRP1
Kai Zurhove et al. Sci Signal. 2008.
Abstract
Inflammation is a potentially self-destructive process that needs tight control. We have identified a nuclear signaling mechanism through which the low-density lipoprotein receptor-related protein 1 (LRP1) limits transcription of lipopolysaccharide (LPS)-inducible genes. LPS increases the proteolytic processing of the ectodomain of LRP1, which results in the gamma-secretase-dependent release of the LRP1 intracellular domain (ICD) from the plasma membrane and its translocation to the nucleus, where it binds to and represses the interferon-gamma promoter. Basal transcription of LPS target genes and LPS-induced secretion of proinflammatory cytokines are increased in the absence of LRP1. The interaction between LRP1-ICD and interferon regulatory factor 3 (IRF-3) promotes the nuclear export and proteasomal degradation of IRF-3. Feedback inhibition of the inflammatory response through intramembranous processing of LRP1 thus defines a physiological role for gamma-secretase.
Figures
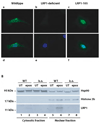
Figure 1. Nuclear localization of the free LRP1 ICD
A) Wildtype (a and d), LRP1-deficient (b and e), and murine embryonic fibroblasts stably expressing the free LRP1 ICD (LRP1-105, c and f) were analyzed by immunocytochemistry with an antibody directed against the LRP1 C-terminus after treatment with 1 µM epoxomicin for 9h. Nuclear counterstaining was done by DAPI (d–f). A representative experiment of n=5 is shown. B) Nuclear and cytosolic extracts were prepared from wildtype (WT) and LRP1-deficient (k.o.) MEF after treatment with 1 µM epoxomicin or no treatment for 12h and were subsequently analyzed by Western Blotting with the C-terminal LRP1-antibody, a nuclear marker (α-Histone 2b), and a cytosolic marker (α-Hsp90). A representative experiment of n=5 is shown.
Figure 2. LPS enhances proteolytical processing of LRP1
A) Wildtype primary macrophages were pretreated with the γ-secretase inhibitor DAPT at a final concentration of 10 µM for 2 h. Then cells were treated with 1 µg/ml LPS or left untreated for 9 h and cell membranes were prepared and analyzed by Western Blotting with the C-terminal LRP1 antibody. A representative experiment of n=4 is shown. B) Schematic representation of LPS-induced signaling pathways. Binding of LPS to its receptor complex leads to the activation of MyD88 (and Mal)-dependent signaling that results in the early activation of NFκB, and of MyD88-independent (TRIF and TRAM-dependent) pathways that lead to the activation of IRF-3 and late NFκB activation. TRIF – TIR-domain containing adaptor molecule, TRAM – TRIF-related adapter molecule, MyD88 – myeloid differentiation marker 88, MAL – MyD88-like, TRAF6 – tumor necrosis factor receptor-associated factor, TBK1 – TANK-binding protein kinase, NEMO – NFκB essential modulator, IKK – IκB kinase.
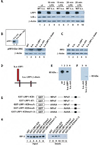
Figure 3. LRP1 modulates LPS-activated signaling through direct interaction with IRF-3
A) Whole cell lysates from wildtype and LRP1-deficient fibroblasts were prepared after treatment with 10 µg/ml LPS for the times indicated and from untreated controls. Lysates were analyzed by Western Blotting and staining with an anti-IκBα antibody. Anti-β-actin was used as a loading control. For quantification of Western Blotting results see supplementary Figure 1A. B) Nuclear extracts were prepared from wildtype and LRP1-deficient fibroblasts as well as from LRP1-deficient cells stably transfected with the LRP1-cDNA (k.o.-LRP1), the empty plasmid vector (k.o.-ctrl.), or the LRP1-βchain (k.o.-LRP1-βchain). The extracts were analyzed by Western Blotting and staining with an anti-phospho-IRF-3 antibody. Staining for β-actin served as loading control. Inset: Analysis of nuclear extracts from LRP1-deficient cells stably transfected with the LRP1-ICD-cDNA (k.o.-LRP1-105) and the empty plasmid vector (k.o.-ctrl.) A representative experiment of n=5 is shown. C) Total IRF-3 levels in the cell lines described under B) were identical as judged by analysis of whole cell lysates by Western Blotting and staining with an IRF-3 antibody. Staining with a β-actin antibody served as loading control. D) Schematic representation of LRP1 and the LRP1 mutants retransfected into the LRP1-deficient fibroblasts. E)/F) Wildtype (wt) and LRP1-deficient (k.o.) MEF and k.o. cells retransfected with LRP1, the LRP1-β-chain, or the empty plasmid vector were analyzed for LRP1 expression by Western Blotting with the C-terminal LRP1 antibody. G) Schematic representation of the GST-LRP1 fusion proteins used in a pull-down assay to detect direct interaction of IRF-3 with LRP1. The LRP ICD was N-terminally fused to a GST tag. In addition a C-terminal LRP1 ICD deletion mutant and constructs with mutations in the first, second, or both NPxY motifs were employed. The black box indicates a putative casein kinase II phosphorylation site in the distal LRP1 ICD. H) GST-LRP1 ICD fusion proteins (see G) were used to pull down IRF-3 from whole cell lysates of MEF. Equal input of fusion proteins was made visible by Ponceau staining of the transfer membrane (see supplementary figure 7). A representative experiment of n=3 is shown.
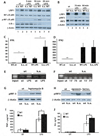
Figure 4. The LRP1 ICD regulates the expression of a subset of LPS-induced genes by direct nuclear signaling and limiting nuclear pIRF-3
A) Whole cell lysates were prepared from wildtype and LRP1-deficient macrophages after treatment with 1 µg/ml LPS for the times indicated and from untreated controls. The lysates were examined as described under 3A). Pooled macrophages from 4 k.o. and 4 wildtype mice were used for each repetition of the experiment (n=3). For quantification of Western Blotting results see supplementary Figure 1B. B) Whole cell lysates were prepared from wildtype (WT) and LRP1-deficient macrophages (k.o.) treated with 1 µg/ml LPS for the times indicated. After Western Blotting antibodies directed against LRP1, pIRF-3, IRF-3 and β-actin were used for staining. Pooled macrophages from 3 k.o. and 3 wildtype mice were used for each repetition of the experiment (n=3). For quantification of Western Blotting results see supplementary Figure 1C. C) Total RNA was prepared from wildtype and LRP1-deficient macrophages either treated with 1 µg/ml LPS or left untreated. Quantitative real-time PCR was used to assess expression levels of interferon γ. Bars represent the mean of 10 independent experiments, error bars depict SEM. Statistical analysis was done using the Student’s t-test. * p<0.05. For each of the 10 experiments one conditional k.o. and one control mouse were used. D) Total RNA was prepared from wildtype and LRP1-deficient macrophages either treated with 1 µg/ml LPS or left untreated. Quantitative real-time PCR was used to assess expression levels of interferon β. Bars represent the mean of 6 independent experiments, error bars depict SEM. Statistical analysis was done using the Student’s t-test. * p<0.01. For each of the 6 experiments one conditional k.o. and one control mouse were used. E) Wildtype and LRP1-deficient macrophages were treated with 1 µg/ml LPS for 15 min. or left untreated. Then DNA-binding proteins were cross-linked to the chromatin and cell lysates were prepared. After shearing of the DNA by sonification an immunoprecipitation with the C-terminal LRP1 antibody was carried out. The precipitate was used in a PCR reaction with primers binding to the interferon γ promoter. A representative experiment of n=3 is shown. For each of the 3 experiments one conditional k.o. and one control mouse were used. F) Wildtype and LRP1-deficient macrophages were treated with 1 µg/ml LPS for the times indicated or left untreated. Chromatin immunoprecipitation was carried out as described above. Primers specific for the interferon β promoter were used in the PCR reaction. A representative experiment of n=3 is shown. For each of the 3 experiments one conditional k.o. and one control mouse were used. G) Wildtype and LRP1-deficient murine embryonic fibroblasts were treated with 4 ng/ml leptomycin B for 19h or were left untreated. Nuclear extracts were prepared and analyzed by immunoblotting with a pIRF-3 antibody. β-actin served as a loading control. A representative experiment of n=3 is shown. H) Wildtype and LRP1-deficient murine embryonic fibroblasts were treated with 1 µM epoxomicin or 10 µM lactacystin for 19h or were left untreated. Nuclear extracts were prepared and analyzed by immunoblotting with a pIRF-3 antibody. β-actin served as a loading control. I) Wildtype and LRP1-deficient macrophages were treated with 10 ng/ml LPS for 4h or left untreated. Supernatants were then collected and TNF-α concentrations were determined by ELISA. Bars represent the mean of 6 independent experiments, error bars depict SEM. Statistical analysis was done using the Student’s t-test. * p<0.005. For each of the 6 experiments one conditional k.o. and one control mouse were used. J) Wildtype and LRP1-deficient macrophages were treated with 1 µg/ml LPS for 24h or left untreated. Supernatants were then collected and IL-6 concentrations were determined by ELISA. Bars represent the mean of 6 independent experiments, error bars depict SEM. Statistical analysis was done using the Student’s t-test. * p<0.05. For each of the 6 experiments one conditional k.o. and one control mouse were used.
Figure 5. Identification of a subset of LPS-inducible genes that are repressed by the LRP1 ICD
A) Total RNA from wildtype and LRP1-deficient MEF and from LRP1-deficient fibroblasts stably re-transfected with LRP1, the membrane-bound LRP1 ICD (LRP-β-chain), the LRP1 ECD, the free LRP1 ICD (LRP1-105) or the empty plasmid vector was analyzed by quantitative real-time PCR for expression levels of Usp18, Rsad2, Tyki, and Oas1. Bars represent the mean of 6 independent experiments, error bars depict SEM. Statistical analysis was done using the Student’s t-test. * p<0.001. B) Wildtype fibroblasts were treated with 2 µg/ml LPS for 3h or were left untreated. RNA was prepared and examined by real-time PCR for expression of Usp18, Rsad2, Tyki, and Oas1. ** p<0.05, n=5.
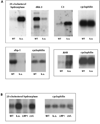
Figure 6. LRP1 is involved in multiple signaling pathways
A) RNA samples from wildtype and LRP1-deficient MEF were analyzed by Northern Blotting with cDNA probes for the genes indicated. Cyclophilin served as a loading control. A representative experiment of n=3 is shown. Dkk-3 – dickkopf-3, C3 – complement factor 3, sfrp-1 – secreted frizzled-related protein-1, AHR – aryl hydrocarbon receptor. B) RNA samples from wildtype and LRP1-deficient MEF and from LRP1-deficient fibroblasts stably re-transfected with an expression plasmid for LRP1 (control: empty plasmid vector) were examined for 25-cholesterol-hydroxylase expression. A representative experiment of n=3 is shown.
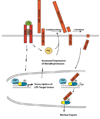
Figure 7. Proposed model for the negative feedback regulation of LPS-induced inflammatory signaling by γ-secretase-dependent LRP1 ICD generation
Expression of a subset of LPS-induced genes is activated by IRF-3. Activation of IRF-3 and other LPS-induced signaling pathways leads to increased expression of metalloproteases. In addition, LPS-dependent PKC-activation also occurs (55). The proteolytical processing of LRP1 is therefore augmented. Increased shedding provides more substrate for the γ-secretase-mediated cleavage step that leads to LRP1 ICD release. The LRP1 ICD interacts with IRF-3 and displaces it from its binding to CBP/p300, thereby unmasking its nuclear export signal (yellow) and facilitating its nuclear export. Subsequently, IRF-3 target gene expression is reduced.
Similar articles
- Inhibition of ADAM10 promotes the clearance of Aβ across the BBB by reducing LRP1 ectodomain shedding.
Shackleton B, Crawford F, Bachmeier C. Shackleton B, et al. Fluids Barriers CNS. 2016 Aug 8;13(1):14. doi: 10.1186/s12987-016-0038-x. Fluids Barriers CNS. 2016. PMID: 27503326 Free PMC article. - Differential glycosylation regulates processing of lipoprotein receptors by gamma-secretase.
May P, Bock HH, Nimpf J, Herz J. May P, et al. J Biol Chem. 2003 Sep 26;278(39):37386-92. doi: 10.1074/jbc.M305858200. Epub 2003 Jul 18. J Biol Chem. 2003. PMID: 12871934 - Regulated intramembrane proteolysis of the low-density lipoprotein receptor-related protein mediates ischemic cell death.
Polavarapu R, An J, Zhang C, Yepes M. Polavarapu R, et al. Am J Pathol. 2008 May;172(5):1355-62. doi: 10.2353/ajpath.2008.070975. Epub 2008 Apr 10. Am J Pathol. 2008. PMID: 18403601 Free PMC article. - LDL receptor-related protein-1: a regulator of inflammation in atherosclerosis, cancer, and injury to the nervous system.
Gonias SL, Campana WM. Gonias SL, et al. Am J Pathol. 2014 Jan;184(1):18-27. doi: 10.1016/j.ajpath.2013.08.029. Epub 2013 Oct 12. Am J Pathol. 2014. PMID: 24128688 Free PMC article. Review. - gamma-Secretase: a multifaceted regulator of angiogenesis.
Boulton ME, Cai J, Grant MB. Boulton ME, et al. J Cell Mol Med. 2008 Jun;12(3):781-95. doi: 10.1111/j.1582-4934.2008.00274.x. Epub 2008 Feb 8. J Cell Mol Med. 2008. PMID: 18266961 Free PMC article. Review.
Cited by
- Genome wide CRISPR screen for Pasteurella multocida toxin (PMT) binding proteins reveals LDL Receptor Related Protein 1 (LRP1) as crucial cellular receptor.
Schoellkopf J, Mueller T, Hippchen L, Mueller T, Reuten R, Backofen R, Orth J, Schmidt G. Schoellkopf J, et al. PLoS Pathog. 2022 Dec 14;18(12):e1010781. doi: 10.1371/journal.ppat.1010781. eCollection 2022 Dec. PLoS Pathog. 2022. PMID: 36516199 Free PMC article. - Low-density lipoprotein receptor-related protein-1: role in the regulation of vascular integrity.
Strickland DK, Au DT, Cunfer P, Muratoglu SC. Strickland DK, et al. Arterioscler Thromb Vasc Biol. 2014 Mar;34(3):487-98. doi: 10.1161/ATVBAHA.113.301924. Epub 2014 Feb 6. Arterioscler Thromb Vasc Biol. 2014. PMID: 24504736 Free PMC article. Review. - Role of LRP1 in the pathogenesis of Alzheimer's disease: evidence from clinical and preclinical studies.
Shinohara M, Tachibana M, Kanekiyo T, Bu G. Shinohara M, et al. J Lipid Res. 2017 Jul;58(7):1267-1281. doi: 10.1194/jlr.R075796. Epub 2017 Apr 4. J Lipid Res. 2017. PMID: 28381441 Free PMC article. Review. - Low-Density Lipoprotein Receptor-Related Protein-1 Signaling in Angiogenesis.
Mao H, Xie L, Pi X. Mao H, et al. Front Cardiovasc Med. 2017 May 22;4:34. doi: 10.3389/fcvm.2017.00034. eCollection 2017. Front Cardiovasc Med. 2017. PMID: 28589128 Free PMC article. Review. - Identification of the low density lipoprotein (LDL) receptor-related protein-1 interactome in central nervous system myelin suggests a role in the clearance of necrotic cell debris.
Fernandez-Castaneda A, Arandjelovic S, Stiles TL, Schlobach RK, Mowen KA, Gonias SL, Gaultier A. Fernandez-Castaneda A, et al. J Biol Chem. 2013 Feb 15;288(7):4538-48. doi: 10.1074/jbc.M112.384693. Epub 2012 Dec 21. J Biol Chem. 2013. PMID: 23264627 Free PMC article.
References
- Parks AL, Curtis D. Presenilin diversifies its portfolio. Trends Genet. 2007;23:140–150. - PubMed
- Ehebauer M, Hayward P, Martinez-Arias A. Notch signaling pathway. Sci STKE. 2006;2006:cm7. - PubMed
- Ni CY, Murphy MP, Golde TE, Carpenter G. gamma -Secretase cleavage and nuclear localization of ErbB-4 receptor tyrosine kinase. Science. 2001;294:2179–2181. - PubMed
- Sardi SP, Murtie J, Koirala S, Patten BA, Corfas G. Presenilin-dependent ErbB4 nuclear signaling regulates the timing of astrogenesis in the developing brain. Cell. 2006;127:185–197. - PubMed
MeSH terms
Substances
Grants and funding
- R01 HL063762-02/HL/NHLBI NIH HHS/United States
- R01 NS043408/NS/NINDS NIH HHS/United States
- P01 HL020948/HL/NHLBI NIH HHS/United States
- R01 HL063762-07/HL/NHLBI NIH HHS/United States
- P01 HL020948-250015/HL/NHLBI NIH HHS/United States
- R01 HL063762-06/HL/NHLBI NIH HHS/United States
- P01 HL020948-310005/HL/NHLBI NIH HHS/United States
- R01 HL063762-01/HL/NHLBI NIH HHS/United States
- R01 NS043408-05/NS/NINDS NIH HHS/United States
- P01 HL020948-240015/HL/NHLBI NIH HHS/United States
- R01 HL063762-04/HL/NHLBI NIH HHS/United States
- R01 HL063762-09/HL/NHLBI NIH HHS/United States
- R01 NS043408-01/NS/NINDS NIH HHS/United States
- R37 HL063762/HL/NHLBI NIH HHS/United States
- R01 HL063762-10/HL/NHLBI NIH HHS/United States
- R01 HL063762-05/HL/NHLBI NIH HHS/United States
- P01 HL020948-210015/HL/NHLBI NIH HHS/United States
- R01 NS043408-03/NS/NINDS NIH HHS/United States
- R01 NS043408-04/NS/NINDS NIH HHS/United States
- R01 HL063762/HL/NHLBI NIH HHS/United States
- P01 HL020948-320005/HL/NHLBI NIH HHS/United States
- R01 HL063762-03/HL/NHLBI NIH HHS/United States
- R01 HL063762-08/HL/NHLBI NIH HHS/United States
- P01 HL020948-230015/HL/NHLBI NIH HHS/United States
- R01 NS043408-02/NS/NINDS NIH HHS/United States
- P01 HL020948-220015/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous