Highly sensitive feature detection for high resolution LC/MS - PubMed (original) (raw)
Highly sensitive feature detection for high resolution LC/MS
Ralf Tautenhahn et al. BMC Bioinformatics. 2008.
Abstract
Background: Liquid chromatography coupled to mass spectrometry (LC/MS) is an important analytical technology for e.g. metabolomics experiments. Determining the boundaries, centres and intensities of the two-dimensional signals in the LC/MS raw data is called feature detection. For the subsequent analysis of complex samples such as plant extracts, which may contain hundreds of compounds, corresponding to thousands of features -- a reliable feature detection is mandatory.
Results: We developed a new feature detection algorithm centWave for high-resolution LC/MS data sets, which collects regions of interest (partial mass traces) in the raw-data, and applies continuous wavelet transformation and optionally Gauss-fitting in the chromatographic domain. We evaluated our feature detection algorithm on dilution series and mixtures of seed and leaf extracts, and estimated recall, precision and F-score of seed and leaf specific features in two experiments of different complexity.
Conclusion: The new feature detection algorithm meets the requirements of current metabolomics experiments. centWave can detect close-by and partially overlapping features and has the highest overall recall and precision values compared to the other algorithms, matchedFilter (the original algorithm of XCMS) and the centroidPicker from MZmine. The centWave algorithm was integrated into the Bioconductor R-package XCMS and is available from (http://www.bioconductor.org/).
Figures
Figure 1
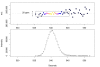
Mass trace and chromatographic peak of Biochanin A [M + _H_]+ mass signal. The upper panel shows the mass trace of the biochanin A [M + _H_]+ mass signal across 10 seconds with colour-coded intensities. The corresponding chromatographic peak is shown below.
Figure 2
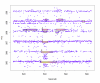
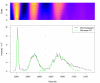
Region Of Interest (ROI) detection. Raw data in the chromatographic and m/z region around the [M + _H_]+ mass signal (1) of biochanin A. In addition to the three isotopic peaks (2–4) other mass signals are marked as ROIs.
Figure 3
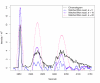
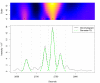
Matched filter effects, example region 1. HPLC/ESI-QTOF-MS of a A. thaliana leaf extract. Extracted ion chromatogram (277.213 – 277.221 m/z) and matched filter results using second derivative Gaussian with different filter widths. Negative filter values were omitted.
Figure 4
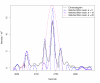
Matched filter effects, example region 2. HPLC/ESI-QTOF-MS of a A. thaliana leaf extract. Extracted ion chromatogram (967.53–967.56 m/z, same sample that was used for Figure 3) and matched filter results using second derivative Gaussian with different filter widths. Negative filter values were clipped.
Figure 5
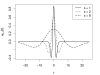
Mexican Hat Wavelet. Mexican hat wavelet at different scales.
Figure 6
centWave results for example region 1. centWave results for example region 1. The lower part shows the same extracted ion chromatogram (277.213–277.221 m/z) as in Figure 3 and the detected chromatographic peaks from the centWave algorithm as Gaussian fits. The upper part shows the CWT coefficients on the different scales. A cross marks the scale where the peak was optimally localised. The vertical grey lines show the peak borders which were estimated from the coefficients of this scale.
Figure 7
centWave results for example region 2. centWave results for example region 2. The lower part shows the same extracted ion chromatogram (967.53–967.56 m/z) as in Figure 4 and the detected chromatographic peaks from the centWave algorithm as Gaussian fits. The upper part shows the CWT coefficients on the different scales. A cross marks the scale where the peak was optimally localised. The vertical grey lines show the peak borders which were estimated from the coefficients of this scale.
Figure 8
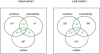
Venn Diagrams of Detected Features. Venn Diagrams showing the number of features in seed and leaf extracts that were found by the three different algorithms. Only the overlapping (green coloured) subsets were used as ground truth.
Figure 9
F-score values for Experiment 1 & 2. F-score (combined measure of recall and precision, calculated from the ground truth features) for dilution series of the seed and leaf extract (left-most and middle part) and for mixtures of the seed and leaf extract (right-most part of the figure). Detected features that match the respective ground truth features were counted als true positives, while all other features returned were considered as false positives. Higher F-score values represent better feature detection performance.
Figure 10
F-score values for Experiment 1 & 2 (alternative parameter settings). F-score (combined measure of recall and precision, calculated from the ground truth features) for dilution series of the seed and leaf extract (left-most and middle part) and for mixtures of the seed and leaf extract (right-most part of the figure). Detected features that match the respective ground truth features were counted als true positives, while all other features returned were considered as false positives. Higher F-score values represent better feature detection performance. Alternative parameter settings were used (see Additional file 4).
Similar articles
- GridMass: a fast two-dimensional feature detection method for LC/MS.
Treviño V, Yañez-Garza IL, Rodriguez-López CE, Urrea-López R, Garza-Rodriguez ML, Barrera-Saldaña HA, Tamez-Peña JG, Winkler R, Díaz de-la-Garza RI. Treviño V, et al. J Mass Spectrom. 2015 Jan;50(1):165-74. doi: 10.1002/jms.3512. J Mass Spectrom. 2015. PMID: 25601689 - Critical assessment of alignment procedures for LC-MS proteomics and metabolomics measurements.
Lange E, Tautenhahn R, Neumann S, Gröpl C. Lange E, et al. BMC Bioinformatics. 2008 Sep 15;9:375. doi: 10.1186/1471-2105-9-375. BMC Bioinformatics. 2008. PMID: 18793413 Free PMC article. - [A review and research prospects on the application of the XCMS mass-spectrometry data-processing software in the environmental science field].
Yang C, Zhang A, Gao ZQ, Su GY. Yang C, et al. Se Pu. 2025 Jun;43(6):585-593. doi: 10.3724/SP.J.1123.2025.01019. Se Pu. 2025. PMID: 40394738 Free PMC article. Review. Chinese. - Role of liquid chromatography-high-resolution mass spectrometry (LC-HR/MS) in clinical toxicology.
Wu AH, Gerona R, Armenian P, French D, Petrie M, Lynch KL. Wu AH, et al. Clin Toxicol (Phila). 2012 Sep;50(8):733-42. doi: 10.3109/15563650.2012.713108. Epub 2012 Aug 13. Clin Toxicol (Phila). 2012. PMID: 22888997 Review.
Cited by
- eMZed: an open source framework in Python for rapid and interactive development of LC/MS data analysis workflows.
Kiefer P, Schmitt U, Vorholt JA. Kiefer P, et al. Bioinformatics. 2013 Apr 1;29(7):963-4. doi: 10.1093/bioinformatics/btt080. Epub 2013 Feb 15. Bioinformatics. 2013. PMID: 23418185 Free PMC article. - Natural variation of root exudates in Arabidopsis thaliana-linking metabolomic and genomic data.
Mönchgesang S, Strehmel N, Schmidt S, Westphal L, Taruttis F, Müller E, Herklotz S, Neumann S, Scheel D. Mönchgesang S, et al. Sci Rep. 2016 Jul 1;6:29033. doi: 10.1038/srep29033. Sci Rep. 2016. PMID: 27363486 Free PMC article. - Pure Ion Chromatograms Combined with Advanced Machine Learning Methods Improve Accuracy of Discriminant Models in LC-MS-Based Untargeted Metabolomics.
Tian M, Lin Z, Wang X, Yang J, Zhao W, Lu H, Zhang Z, Chen Y. Tian M, et al. Molecules. 2021 May 5;26(9):2715. doi: 10.3390/molecules26092715. Molecules. 2021. PMID: 34063107 Free PMC article. - A live yeast supplementation to gestating ewes improves bioactive molecule composition in colostrum with no impact on its bacterial composition and beneficially affects immune status of the offspring.
Dunière L, Renaud JB, Steele MA, Achard CS, Forano E, Chaucheyras-Durand F. Dunière L, et al. J Nutr Sci. 2022 Feb 7;11:e5. doi: 10.1017/jns.2022.3. eCollection 2022. J Nutr Sci. 2022. PMID: 35291274 Free PMC article. - The nPYc-Toolbox, a Python module for the pre-processing, quality-control and analysis of metabolic profiling datasets.
Sands CJ, Wolfer AM, Correia GDS, Sadawi N, Ahmed A, Jiménez B, Lewis MR, Glen RC, Nicholson JK, Pearce JTM. Sands CJ, et al. Bioinformatics. 2019 Dec 15;35(24):5359-5360. doi: 10.1093/bioinformatics/btz566. Bioinformatics. 2019. PMID: 31350543 Free PMC article.
References
- Oliver S, Winson M, Kell D, Baganz F. Systematic functional analysis of the yeast genome. Trends Biotechnol. 1998;16:373–378. - PubMed
- Fiehn O, Kopka J, Dörmann P, Altmann T, Trethewey R, Willmitzer L. Metabolite profiling for plant functional genomics. Nature Biotechnology. 2000;18:115. - PubMed
- Dunn WB. Current trends and future requirements for the mass spectrometric investigation of microbial, mammalian and plant metabolomes. Physical Biology. 2008;5:24. http://stacks.iop.org/1478-3975/5/011001 - PubMed
- Roepenack-Lahaye Ev, Degenkolb T, Zerjeski M, Franz M, Roth U, Wessjohann L, Schmidt J, Scheel D, Clemens S. Profiling of Arabidopsis Secondary Metabolites by Capillary Liquid Chromatography Coupled to Electrospray Ionization Quadrupole Time-of-Flight Mass Spectrometry. Plant Physiology. 2004;134:548–559. - PMC - PubMed
- Böttcher C, Roepenack-Lahaye Ev, Schmidt J, Schmotz C, Neumann S, Scheel D, Clemens S. Metabolome Analysis of Biosynthetic Mutants Reveals Diversity of Metabolic Changes and Allows Identification of a Large Number of New Compounds in Arabidopsis thaliana. Plant Physiol. 2008. p. 108.117754.http://www.plantphysiol.org/cgi/content/abstract/pp.108.117754v1 - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases