In vitro and in vivo analysis of B-Myb in basal-like breast cancer - PubMed (original) (raw)
In vitro and in vivo analysis of B-Myb in basal-like breast cancer
A R Thorner et al. Oncogene. 2009.
Abstract
A defining feature of basal-like breast cancer, a breast cancer subtype with poor clinical prognosis, is the high expression of 'proliferation signature' genes. We identified B-Myb, a MYB family transcription factor that is often amplified and overexpressed in many tumor types, as being highly expressed in the proliferation signature. However, the roles of B-Myb in disease progression, and its mammary-specific transcriptional targets, are poorly understood. Here, we showed that B-Myb expression is a significant predictor of survival and pathological complete response to neoadjuvant chemotherapy in breast cancer patients. We also identified a significant association between the G/G genotype of a nonsynonymous B-Myb germline variant (rs2070235, S427G) and an increased risk of basal-like breast cancer [OR 2.0, 95% CI (1.1-3.8)]. In immortalized, human mammary epithelial cell lines, but not in basal-like tumor lines, cells ectopically expressing wild-type B-Myb or the S427G variant showed increased sensitivity to two DNA topoisomerase IIalpha inhibitors, but not to other chemotherapeutics. In addition, microarray analyses identified many G2/M genes as being induced in B-Myb overexpressing cells. These results confirm that B-Myb is involved in cell cycle control, and that its dysregulation may contribute to increased sensitivity to a specific class of chemotherapeutic agents. These data provide insight into the influence of B-Myb in human breast cancer, which is of potential clinical importance for determining disease risk and for guiding treatment.
Figures
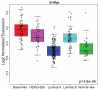
Figure 1. B-Myb expression across breast cancer subtypes
The NKI breast tumor microarray dataset (n=295) was classified into the five intrinsic subtypes and box plots used to visualize B-Myb expression according to breast cancer subtypes. Statistical significance was calculated by ANOVA.
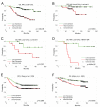
Figure 2. High expression of B-Myb correlates with poor outcome
Kaplan-Meier survival analyses based on B-Myb expression values rank ordered into halves (low/high). (A-D) Overall survival (OS) of locally treated NKI tumor samples: (A) All subtypes combined (n=165), (B) Luminal A (n=72), (C) Luminal B (n=26), (D) HER2+/ER− (n=21). (E) RFS, Wang et al., 2005 (n=286), a locally treated, lymph-node-negative tumor microarray dataset. (F) Miller et al., 2005 breast tumor microarray dataset (n=234).
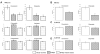
Figure 3. Drug sensitivities in B-Myb overexpressing cell lines
IC50 doses (72h) of chemotherapy on cell lines stably expressing vector control, B-Myb, or B-Myb S427G variant. Each MTT experiment was performed in triplicate and error bars represent 95% confidence intervals (*p<0.001 relative to vector control). (A) _hTERT_-immortalized HMEC line HME-CC. (B) _hTERT_-immortalized HMEC line ME16C. (C) Basal-like tumor derived cell line SUM102 and (D) SUM149.
Figure 4. Enrichment of G2/M phase genes in doxorubicin-treated B-Myb overexpressing HME-CC cells
Significance Analysis of Microarray was used to identify 217 significant genes whose high expression was present in B-Myb overexpressing cells. These genes were then assigned to a specific phase of the cell cycle by comparing them to Whitfield et al., 2002, which identified 101/217 genes as being specifically induced during the cell cycle. The graph shows to which phase of the cell cycle these 101 genes mapped.
Figure 5. Cell cycle profile of HME-CC cells stably expressing B-Myb and treated with doxorubicin
Cell cultures were treated with a range of doses of doxorubicin for 48 hours followed by propidium iodide DNA content analysis. Percentage of cells in (A) G1 phase and (B) G2/M were calculated by gating based on DNA content. Error bars indicate standard deviations between three independent experiments.
References
- Bergamaschi A, Kim YH, Wang P, Sørlie T, Hernandez-Boussard T, Lonning PE, et al. Distinct patterns of DNA copy number alteration are associated with different clinicopathological features and gene-expression subtypes of breast cancer. Genes, Chromosomes and Cancer. 2006;45:1033–1040. - PubMed
- Carey LA, Dees EC, Sawyer L, Gatti L, Moore DT, Collichio F, et al. The Triple Negative Paradox : Primary Tumor Chemosensitivity of Breast Cancer Subtypes. Clin Cancer Res. 2007;13:2329–2334. - PubMed
- Carey LA, Perou CM, Livasy CA, Dressler LG, Cowan D, Conway K, et al. Race, Breast Cancer Subtypes, and Survival in the Carolina Breast Cancer Study. JAMA. 2006;295:2492–2502. - PubMed
- Chin K, DeVries S, Fridlyand J, Spellman PT, Roydasgupta R, Kuo W-L, et al. Genomic and transcriptional aberrations linked to breast cancer pathophysiologies. Cancer Cell. 2006;10:529–541. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P50 CA058223/CA/NCI NIH HHS/United States
- R01 CA101227/CA/NCI NIH HHS/United States
- R01-CA-101227-01/CA/NCI NIH HHS/United States
- P50-CA58223-09A1/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases