SRA-domain proteins required for DRM2-mediated de novo DNA methylation - PubMed (original) (raw)
SRA-domain proteins required for DRM2-mediated de novo DNA methylation
Lianna M Johnson et al. PLoS Genet. 2008 Nov.
Abstract
De novo DNA methylation and the maintenance of DNA methylation in asymmetrical sequence contexts is catalyzed by homologous proteins in plants (DRM2) and animals (DNMT3a/b). In plants, targeting of DRM2 depends on small interfering RNAs (siRNAs), although the molecular details are still unclear. Here, we show that two SRA-domain proteins (SUVH9 and SUVH2) are also essential for DRM2-mediated de novo and maintenance DNA methylation in Arabidopsis thaliana. At some loci, SUVH9 and SUVH2 act redundantly, while at other loci only SUVH2 is required, and this locus specificity correlates with the differing DNA-binding affinity of the SRA domains within SUVH9 and SUVH2. Specifically, SUVH9 preferentially binds methylated asymmetric sites, while SUVH2 preferentially binds methylated CG sites. The suvh9 and suvh2 mutations do not eliminate siRNAs, suggesting a role for SUVH9 and SUVH2 late in the RNA-directed DNA methylation pathway. With these new results, it is clear that SRA-domain proteins are involved in each of the three pathways leading to DNA methylation in Arabidopsis.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
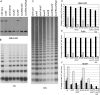
Figure 1. suvh9 suvh2 kyp triple mutants show a similar phenotype as drm1 drm2 cmt3 triple mutants.
A. Curled-leaf phenotype of young seedlings. Wild-type line is Columbia accession and all mutants are homozygous for T-DNA insertions in the Columbia background. The developmental phenotype appears in the first generation. B. Short stature phenotype observed in suvh9 suvh2 kyp plants. C. Quantitative RT-PCR of SDC gene expression compared to ACTIN plotted on log scale. D. Bisulfite sequence results of the SDC tandem repeats region. Between 13–21 independent clones were sequenced and the results are shown as the average number of methyl cytosines per clone. See Figure S2 for methylation expressed as percentage of methyl cytosine and Figure S3 for alignments. Black bar represents CG, gray CHG, and white is CHH methylation.
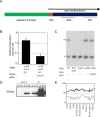
Figure 2. SUVH9 and/or SUVH2 are required for maintenance of non-CG methylation.
A. Southern blot of MspI digested DNA using MEA-ISR probe. Upper band represents methylated DNA and lower band represents unmethylated DNA. B. Same blot as in A reprobed with 5S DNA probe. C. Similar blot probed with CEN180 repeats. D. Bisulfite sequencing results of the MEA-ISR region expressed as the average number of methyl cytosines per clone (approximately 20 clones analyzed). Black (mCG), gray (mCHG), white (mCHH). E. Bisulfite results at FWA. F. Bisulfite results at AtSN1. See Figure S3 for alignments of all bisulfite sequence data.
Figure 3. Epitope tagged SUVH9 and SUVH2 complement the suvh9 and suvh2 mutations.
A. Diagram of epitope-tagged SUVH9 and SUVH2. The TAG is either 3xHA or 9xMyc as described in Material and Methods. B. Quantitative RT-PCR of SDC gene relative to ACTIN in suvh9 suvh2 kyp untransformed and transformed with myc-tagged SUVH9. C. Complementation of suvh2 was tested by MEA-ISR southern blot using MspI digested DNA isolated from Columbia (wt), drm1drm2, suvh2, suvh9, and suvh2 transformed with HA-tagged SUVH2. D. Western blot of HA- tagged SUVH2 and SUVH9 under control of their endogenous promoters in transgenic plants. Input refers to leaf extract before immunoprecipitation, IP after immunoprecipitation. E. Expression levels of SUVH9 and SUVH2 from different tissues as measured on microarrays.
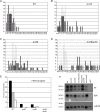
Figure 4. SUVH9 and SUVH2 are required for establishment of DNA methylation and silencing at FWA.
A–D. Flowering time distributions shown as total leaf number of T1 populations of untransformed (light gray) or FWA transformed (black) plants of the indicated genotype. X-axes show total number of leaves at flowering time, and the Y-axes show the percentage of plants with the given leaf number. E. Bisulfite sequencing results of the FWA transgenes of late-flowering T1 plants. Black, CG methylation; gray, CHG methylation; white, CHH methylation. F. siRNA accumulation in various mutant backgrounds. MicroRNA 159 (mi159) was used as a loading control for SDC (siRNAs hybridizing to the tandem repeats) and 5S siRNAs.
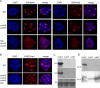
Figure 5. SUVH9 and SUVH2 may be inactive for histone methyltransferase activity.
A–B. Immunofluorescence of H3K9me1, H3K9me2 and H3K27me1 compared to DAPI in nuclei isolated from Columbia (wt), suvh9 suvh2, kyp suvh5 suvh6 and suvh9 suvh2 suvh6 kyp. C. Histone methylation assays using GST fusion proteins and either calf thymus histones (top panel, autoradiogram) or Arabidopsis nucleosomes (middle panel, autoradiogram) as substrates. Calf thymus histones are shown in bottom panel (protein stain). D. 3H-AdoMet crosslinking to GST fusion proteins. Top panel autoradiogram, bottom panel stained proteins.
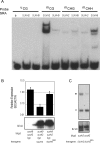
Figure 6. SRA domains of SUVH9 and SUVH2 bind to methylated CHH and CG sequences, respectively.
A. Mobility shift assays using either GST-SUVH2-SRA (SUVH2) or GST-SUVH9-SRA (SUVH9) and either unmethylated CG oligonucleotide (uCG), methylated CG (mCG), methylated CHG (mCHG) or methylated CHH (mCHH) as probe. B. Upper panel: Quantitative RT-PCR of the SDC gene relative to ACTIN was measured in suvh9 suvh2 kyp lines containing the following stable transgenes: myc-tagged SUVH9 (SUVH9) or myc-tagged SUVH9-SRA mutant (S252F; SUVH9SRA). Lower panel: Western blot of transgenic plant extracts probed with myc antibody. C. Upper panel: Complementation of suvh2 was tested by MEA-ISR Southern blot using MspI digested DNA isolated from suvh2 lines containing the following stable transgenes: HA-tagged SUVH2 or HA-tagged SUVH2-SRA mutant (E262K; SUVH2SRA). Lower Panel: Western blot of immunoprecipitated protein from suvh2 transgenic plants using HA antibody as probe.
Similar articles
- SRA- and SET-domain-containing proteins link RNA polymerase V occupancy to DNA methylation.
Johnson LM, Du J, Hale CJ, Bischof S, Feng S, Chodavarapu RK, Zhong X, Marson G, Pellegrini M, Segal DJ, Patel DJ, Jacobsen SE. Johnson LM, et al. Nature. 2014 Mar 6;507(7490):124-128. doi: 10.1038/nature12931. Epub 2014 Jan 22. Nature. 2014. PMID: 24463519 Free PMC article. - The SET domain proteins SUVH2 and SUVH9 are required for Pol V occupancy at RNA-directed DNA methylation loci.
Liu ZW, Shao CR, Zhang CJ, Zhou JX, Zhang SW, Li L, Chen S, Huang HW, Cai T, He XJ. Liu ZW, et al. PLoS Genet. 2014 Jan;10(1):e1003948. doi: 10.1371/journal.pgen.1003948. Epub 2014 Jan 22. PLoS Genet. 2014. PMID: 24465213 Free PMC article. - Developmentally non-redundant SET domain proteins SUVH2 and SUVH9 are required for transcriptional gene silencing in Arabidopsis thaliana.
Kuhlmann M, Mette MF. Kuhlmann M, et al. Plant Mol Biol. 2012 Aug;79(6):623-33. doi: 10.1007/s11103-012-9934-x. Epub 2012 Jun 6. Plant Mol Biol. 2012. PMID: 22669745 Free PMC article. - Heterochromatin proteins and the control of heterochromatic gene silencing in Arabidopsis.
Fischer A, Hofmann I, Naumann K, Reuter G. Fischer A, et al. J Plant Physiol. 2006 Feb;163(3):358-68. doi: 10.1016/j.jplph.2005.10.015. Epub 2005 Dec 27. J Plant Physiol. 2006. PMID: 16384625 Review. - Specifications of Targeting Heterochromatin Modifications in Plants.
Wendte JM, Schmitz RJ. Wendte JM, et al. Mol Plant. 2018 Mar 5;11(3):381-387. doi: 10.1016/j.molp.2017.10.002. Epub 2017 Oct 13. Mol Plant. 2018. PMID: 29032247 Review.
Cited by
- An RdDM-independent function of Pol V transcripts in gene regulation and plant defence.
Yuan Y, Liu Y, Han L, Li Y, Qi Y. Yuan Y, et al. Nat Plants. 2024 Oct;10(10):1562-1575. doi: 10.1038/s41477-024-01774-0. Epub 2024 Aug 26. Nat Plants. 2024. PMID: 39187700 - Regulatory mechanism of heat-active retrotransposons by the SET domain protein SUVH2.
Niu X, Ge Z, Ito H. Niu X, et al. Front Plant Sci. 2024 Feb 8;15:1355626. doi: 10.3389/fpls.2024.1355626. eCollection 2024. Front Plant Sci. 2024. PMID: 38390294 Free PMC article. - The MBD-ACD DNA methylation reader complex recruits MICRORCHIDIA6 to regulate ribosomal RNA gene expression in Arabidopsis.
Ren Z, Gou R, Zhuo W, Chen Z, Yin X, Cao Y, Wang Y, Mi Y, Liu Y, Wang Y, Fan LM, Deng XW, Qian W. Ren Z, et al. Plant Cell. 2024 Mar 29;36(4):1098-1118. doi: 10.1093/plcell/koad313. Plant Cell. 2024. PMID: 38092516 Free PMC article. - The MOM1 complex recruits the RdDM machinery via MORC6 to establish de novo DNA methylation.
Li Z, Wang M, Zhong Z, Gallego-Bartolomé J, Feng S, Jami-Alahmadi Y, Wang X, Wohlschlegel J, Bischof S, Long JA, Jacobsen SE. Li Z, et al. Nat Commun. 2023 Jul 12;14(1):4135. doi: 10.1038/s41467-023-39751-4. Nat Commun. 2023. PMID: 37438334 Free PMC article. - UV-B-induced modulation of constitutive heterochromatin content in Arabidopsis thaliana.
Johann To Berens P, Golebiewska K, Peter J, Staerck S, Molinier J. Johann To Berens P, et al. Photochem Photobiol Sci. 2023 Sep;22(9):2153-2166. doi: 10.1007/s43630-023-00438-w. Epub 2023 May 25. Photochem Photobiol Sci. 2023. PMID: 37225911
References
- Yoder JA, Walsh CP, Bestor TH. Cytosine methylation and the ecology of intragenomic parasites. Trends Genet. 1997;13:335–340. - PubMed
- Martienssen RA, Colot V. DNA methylation and epigenetic inheritance in plants and filamentous fungi. Science. 2001;293:1070–1074. - PubMed
- Zhang X, Yazaki J, Sundaresan A, Cokus S, Chan SW, et al. Genome-wide high-resolution mapping and functional analysis of DNA methylation in arabidopsis. Cell. 2006;126:1189–1201. - PubMed
- Zilberman D, Gehring M, Tran RK, Ballinger T, Henikoff S. Genome-wide analysis of Arabidopsis thaliana DNA methylation uncovers an interdependence between methylation and transcription. Nat Genet. 2007;39:61–69. - PubMed
- Chan SW, Henderson IR, Jacobsen SE. Gardening the genome: DNA methylation in Arabidopsis thaliana. Nat Rev Genet. 2005;6:351–360. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- GM082045/GM/NIGMS NIH HHS/United States
- R37 GM060398/GM/NIGMS NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- F32 GM082045/GM/NIGMS NIH HHS/United States
- R01 GM060398/GM/NIGMS NIH HHS/United States
- GM60398/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases