Global hypomethylation of genomic DNA in cancer-associated myofibroblasts - PubMed (original) (raw)
. 2008 Dec 1;68(23):9900-8.
doi: 10.1158/0008-5472.CAN-08-1319.
Tamas A Gonda, Mary V Gamble, Martha Salas, Venkatraman Seshan, Shuiping Tu, William S Twaddell, Peter Hegyi, Gyorgy Lazar, Islay Steele, Andrea Varro, Timothy C Wang, Benjamin Tycko
Affiliations
- PMID: 19047171
- PMCID: PMC2670548
- DOI: 10.1158/0008-5472.CAN-08-1319
Global hypomethylation of genomic DNA in cancer-associated myofibroblasts
Le Jiang et al. Cancer Res. 2008.
Abstract
Global hypomethylation has long been recognized as a feature of the malignant epithelial component in human carcinomas. Here we show evidence for this same type of epigenetic alteration in cancer-associated stromal myofibroblasts. We used methylation-sensitive SNP array analysis (MSNP) to profile DNA methylation in early-passage cultures of stromal myofibroblasts isolated from human gastric cancers. The MSNP data indicated widespread hypomethylation in these cells, with rare focal gains of methylation, conclusions that were independently validated by bisulfite sequencing and by a methylation-sensitive cytosine incorporation assay. Immunohistochemistry with anti-5-methylcytosine (anti-5-methyl-C) in a series of gastrectomy specimens showed frequent loss of methylation in nuclei of both the malignant epithelial cells and alpha-smooth muscle actin (ASMA)-positive stromal myofibroblasts of both intestinal-type and diffuse carcinomas. We confirmed this phenomenon and established its onset at the stage of noninvasive dysplastic lesions by immunohistochemistry for anti-5-methyl-C in a transgenic mouse model of multistage gastric carcinogenesis. These findings indicate similar general classes of epigenetic alterations in carcinoma cells and their accompanying reactive stromal cells and add to accumulating evidence for biological differences between normal and cancer-associated myofibroblasts.
Figures
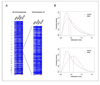
Figure 1. MSNP data showing loss of DNA methylation in cancer-associated myofibroblasts
A, Methylation index values for Class 2 SNPs (Affymetrix 250K StyI arrays) displayed by physical position along the human chromosomes (left panel) and zoomed-in to chromosome 10 (right panel). In each of the 2 cases examined (Case 45, Case 42) the MSNP analysis was a comparison of early passage myofibroblasts isolated from within the polypoid intestinal type carcinoma (CAF) to myofibroblasts isolated from gastric tissue 10 cm away from the carcinoma (NL). Loss of methylation (darker blue color) is seen uniformly across all chromosomes, and the zoomed-in region is simply one example. B, Histograms of methylation index for Class 2 SNPs in the 2 cases. Both cases show a shift in the distribution to lower values in the cancer-associated myofibroblasts, with this loss being most severe in Case 45. The vertical lines indicate the median methylation index for each sample.
Figure 2. Validation of the MSNP data by bisulfite sequencing
A, Bisulfite sequencing of one of the many SNP-tagged loci that showed a reduced methylation index in cancer-associated myofibroblasts from both cases: SNP ID rs10259620; in HOXA9 exon 2. Genomic DNA was bisulfite-treated and then amplified by PCR using primers specific for the converted sequence. The bisulfite PCR products were cloned and multiple clones sequenced (rows). There are a larger number of unmethylated CpG dinucleotides (white circles) in the cancer-associated myofibroblasts (CAF), compared to the myofibroblasts distant from the tumors (NL). B, Results for one of the few SNP-tagged loci (SNP ID rs11079830, HOXB6 proximal promoter region) that showed an increased methylation index in both cases. There are a larger number of methylated CpG dinucleotides (black circles) in the cancer-associated myofibroblasts. The CpG dinucleotides in _Hpa_II sites between the PCR primers are indicated by the vertical arrows. Symbols in the restriction maps are: S, _Sty_I; H, _Hpa_II; horizontal arrows, bisulfite PCR primers.
Figure 3. Validation of loss of DNA methylation in cancer-associated myofibroblasts by a 3HdCTP-Incorporation assay
A, Calibration data from a mixing experiment with in vitro methylated and unmethylated human DNA validating the ability of this assay to quantitate the average CpG methylation of genomic _Hpa_II sites. B, Results with genomic DNA from the early passage gastric myofibroblasts in 5 cases (#42 and #45 are cases represented in the MSNP data while the others are additional cases). There is consistent loss of methylation in the cancer-associated myofibroblasts (CAF) compared to myofibroblasts from non-cancerous gastric mucosa (NL) in all cases, albeit with some variability in the extent of loss from case to case. Mean values and standard deviations of triplicate determinations are shown.
Figure 4. Anti-5-methyl-C IHC showing loss of CpG methylation in myofibroblasts of primary gastric carcinomas
The upper and lower panels show IHC with anti-ASMA and anti-5-methyl-C, respectively. The control non-neoplastic gastric mucosa (Normal) was sampled at a location in the gastrectomy distant from an intestinal type carcinoma. The ASMA-positive spindle-shaped myofibroblasts of the normal muscularis mucosae show strong nuclear staining with anti-5-methyl-C (large arrow), as do the normal epithelial cells (small arrow) and leukocytes (lymphocytes and polymorphonuclear cells; asterisk). The more rare ASMA-positive intraepithelial myofibroblasts also showed uniformly strongly stained in multiple sections examined (example; arrowhead). Sections from within the cancers in 2 cases of intestinal type gastric carcinomas (CA; Case 1 and Case 2) show myofibroblasts (large arrows) and malignant epithelial cells (small arrows) with reduced IHC intensity of 5-methyl-C. As in internal control, the anti-5-methyl-C staining of infiltrating leukocytes remains strong in the neoplastic tissue (asterisks).
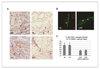
Figure 5. Dual staining to detect nuclear anti-5-methyl-C in ASMA positive cells
A, Dual color IHC (brown DAB: anti-5-methyl-C; blue NBT: anti-ASMA) showing normal and tumor areas from cases of intestinal type (left panels) and diffuse type (right panels) gastric carcinoma, with the mucosa at the uninvolved margin illustrated in the top panels (NL) and the cancerous tissue in the bottom panels (CA). There is an obvious reduction in nuclear 5-methyl-C immunostaining in the cancer-associated myofibroblasts (arrows), while the inflammatory cells surrounding the myofibroblasts have persistently high staining intensity (asterisk). B, Dual color IF from another case of intestinal type gastric cancer. The left panel (NL) shows a confocal scanning microscopy image (100X objective) of non-neoplastic gastric mucosa (from negative surgical margin) showing an intraepithelial ASMA+ (green) cell with strong anti-5-mc staining (red) in the nucleus (arrow). The right panel (CAF) shows a group of intratumoral ASMA+ cells in the intestinal type gastric cancer from the same patient showing decreased nuclear anti-5-methl-C staining (arrows). C, Quantitation of anti-5-methyl-C signal intensity from 10 individual ASMA+ cells in two cases of gastric cancer and adjacent normal tissue. In the intratumoral myofibroblasts from both cases there is a global loss of DNA methylation.
Figure 6. Change in anti-5-methyl-C staining of myofibroblasts in an IL1β transgenic mouse model of progressive gastric dysplasia
Strong anti-5-methyl-C staining is seen for the epithelial cells (small arrows) and ASMA positive myofibroblasts (large arrows) in normal histology IL1β-Tg gastric mucosa. There is an obvious decrease in anti-5-methyl-C staining in both epithelial and ASMA positive cells in the dysplastic gastric mucosa, and a persistent decrease in anti-5-methyl-C staining in epithelial and ASMA positive cells in the carcinoma in situ. As an internal control, there is preserved anti-5-methyl-C staining of infiltrating lymphocytes in both the dysplasia and carcinoma in situ. These IHC findings were consistent in each of two animals examined from each of the 3 categories (normal, dysplasia, CA in situ).
Similar articles
- Folic acid increases global DNA methylation and reduces inflammation to prevent Helicobacter-associated gastric cancer in mice.
Gonda TA, Kim YI, Salas MC, Gamble MV, Shibata W, Muthupalani S, Sohn KJ, Abrams JA, Fox JG, Wang TC, Tycko B. Gonda TA, et al. Gastroenterology. 2012 Apr;142(4):824-833.e7. doi: 10.1053/j.gastro.2011.12.058. Epub 2012 Jan 13. Gastroenterology. 2012. PMID: 22248660 - Stromal cell-derived factor-1 overexpression induces gastric dysplasia through expansion of stromal myofibroblasts and epithelial progenitors.
Shibata W, Ariyama H, Westphalen CB, Worthley DL, Muthupalani S, Asfaha S, Dubeykovskaya Z, Quante M, Fox JG, Wang TC. Shibata W, et al. Gut. 2013 Feb;62(2):192-200. doi: 10.1136/gutjnl-2011-301824. Epub 2012 Feb 23. Gut. 2013. PMID: 22362916 Free PMC article. - Alu and Satα hypomethylation in Helicobacter pylori-infected gastric mucosae.
Yoshida T, Yamashita S, Takamura-Enya T, Niwa T, Ando T, Enomoto S, Maekita T, Nakazawa K, Tatematsu M, Ichinose M, Ushijima T. Yoshida T, et al. Int J Cancer. 2011 Jan 1;128(1):33-9. doi: 10.1002/ijc.25534. Int J Cancer. 2011. PMID: 20602342 - Genetic pathways of two types of gastric cancer.
Tahara E. Tahara E. IARC Sci Publ. 2004;(157):327-49. IARC Sci Publ. 2004. PMID: 15055305 Review. - Gene methylation in gastric cancer.
Qu Y, Dang S, Hou P. Qu Y, et al. Clin Chim Acta. 2013 Sep 23;424:53-65. doi: 10.1016/j.cca.2013.05.002. Epub 2013 May 10. Clin Chim Acta. 2013. PMID: 23669186 Review.
Cited by
- Fibroblast Reprogramming in Gastrointestinal Cancer.
Melissari MT, Chalkidi N, Sarris ME, Koliaraki V. Melissari MT, et al. Front Cell Dev Biol. 2020 Jul 15;8:630. doi: 10.3389/fcell.2020.00630. eCollection 2020. Front Cell Dev Biol. 2020. PMID: 32760726 Free PMC article. Review. - Loss of Y-chromosome does not correlate with age at onset of head and neck carcinoma: a case-control study.
Veiga LC, Bérgamo NA, Reis PP, Kowalski LP, Rogatto SR. Veiga LC, et al. Braz J Med Biol Res. 2012 Feb;45(2):172-8. doi: 10.1590/s0100-879x2012007500004. Epub 2012 Jan 19. Braz J Med Biol Res. 2012. PMID: 22249426 Free PMC article. - Blood leukocyte Alu and LINE-1 methylation and gastric cancer risk in the Shanghai Women's Health Study.
Gao Y, Baccarelli A, Shu XO, Ji BT, Yu K, Tarantini L, Yang G, Li HL, Hou L, Rothman N, Zheng W, Gao YT, Chow WH. Gao Y, et al. Br J Cancer. 2012 Jan 31;106(3):585-91. doi: 10.1038/bjc.2011.562. Epub 2011 Dec 15. Br J Cancer. 2012. PMID: 22173668 Free PMC article. - Epigenetic control of the tumor microenvironment.
Marks DL, Olson RL, Fernandez-Zapico ME. Marks DL, et al. Epigenomics. 2016 Dec;8(12):1671-1687. doi: 10.2217/epi-2016-0110. Epub 2016 Oct 4. Epigenomics. 2016. PMID: 27700179 Free PMC article. Review. - Epigenetics of Bladder Cancer: Where Biomarkers and Therapeutic Targets Meet.
Martinez VG, Munera-Maravilla E, Bernardini A, Rubio C, Suarez-Cabrera C, Segovia C, Lodewijk I, Dueñas M, Martínez-Fernández M, Paramio JM. Martinez VG, et al. Front Genet. 2019 Nov 18;10:1125. doi: 10.3389/fgene.2019.01125. eCollection 2019. Front Genet. 2019. PMID: 31850055 Free PMC article. Review.
References
- Dvorak HF. Tumors: wounds that do not heal. Similarities between tumor stroma generation and wound healing. The New England journal of medicine. 1986;315:1650–1659. - PubMed
- Tsujino T, Seshimo I, Yamamoto H, et al. Stromal myofibroblasts predict disease recurrence for colorectal cancer. Clin Cancer Res. 2007;13:2082–2090. - PubMed
- Orimo A, Weinberg RA. Stromal fibroblasts in cancer: a novel tumor-promoting cell type. Cell cycle (Georgetown, Tex. 2006;5:1597–1601. - PubMed
- Desmouliere A, Guyot C, Gabbiani G. The stroma reaction myofibroblast: a key player in the control of tumor cell behavior. The International journal of developmental biology. 2004;48:509–517. - PubMed
- Mahadevan D, Von Hoff DD. Tumor-stroma interactions in pancreatic ductal adenocarcinoma. Mol Cancer Ther. 2007;6:1186–1197. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases