Newly introduced genomic prophage islands are critical determinants of in vivo competitiveness in the Liverpool Epidemic Strain of Pseudomonas aeruginosa - PubMed (original) (raw)
doi: 10.1101/gr.086082.108. Epub 2008 Dec 1.
Morgan G I Langille, Joanne L Fothergill, Irena Kukavica-Ibrulj, Catherine Paradis-Bleau, François Sanschagrin, Nicholas R Thomson, Geoff L Winsor, Michael A Quail, Nicola Lennard, Alexandra Bignell, Louise Clarke, Kathy Seeger, David Saunders, David Harris, Julian Parkhill, Robert E W Hancock, Fiona S L Brinkman, Roger C Levesque
Affiliations
- PMID: 19047519
- PMCID: PMC2612960
- DOI: 10.1101/gr.086082.108
Newly introduced genomic prophage islands are critical determinants of in vivo competitiveness in the Liverpool Epidemic Strain of Pseudomonas aeruginosa
Craig Winstanley et al. Genome Res. 2009 Jan.
Abstract
Pseudomonas aeruginosa isolates have a highly conserved core genome representing up to 90% of the total genomic sequence with additional variable accessory genes, many of which are found in genomic islands or islets. The identification of the Liverpool Epidemic Strain (LES) in a children's cystic fibrosis (CF) unit in 1996 and its subsequent observation in several centers in the United Kingdom challenged the previous widespread assumption that CF patients acquire only unique strains of P. aeruginosa from the environment. To learn about the forces that shaped the development of this important epidemic strain, the genome of the earliest archived LES isolate, LESB58, was sequenced. The sequence revealed the presence of many large genomic islands, including five prophage clusters, one defective (pyocin) prophage cluster, and five non-phage islands. To determine the role of these clusters, an unbiased signature tagged mutagenesis study was performed, followed by selection in the chronic rat lung infection model. Forty-seven mutants were identified by sequencing, including mutants in several genes known to be involved in Pseudomonas infection. Furthermore, genes from four prophage clusters and one genomic island were identified and in direct competition studies with the parent isolate; four were demonstrated to strongly impact on competitiveness in the chronic rat lung infection model. This strongly indicates that enhanced in vivo competitiveness is a major driver for maintenance and diversifying selection of these genomic prophage genes.
Figures
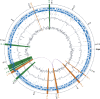
Figure 1.
Circular map of the P. aeruginosa LES genome. Starting from outermost circle going inwards: major (500 kb) and minor tick (100 kb) measurements of the genome with estimated location of the origin; prophage (orange) and genomic islands (green) are highlighted across all tracks; protein coding genes (blue) on plus (outer) and minus strand (inner); tRNAs (green), rRNAs (orange), and all other noncoding RNA genes (purple); signature tagged mutants (black); GC content (outer black line plot) with GC content average (gray line); and GC skew (inner black line plot) were calculated using a 10-kb non-overlapping window. Note that one region of low GC, upstream of the first noted prophage, plus additional smaller regions of low GC, contain ribosomal genes that are commonly known to have a lower GC in genomes. The location of two highly similar genomic regions of length 7.5 kb and 13.5 kb within the prophages are marked with looping purple lines, between their locations on the innermost circle. The identified prophage and GIs are distributed around the genome, but there is one notable cluster of LESGI-1, LESGI-2, and LESGI-3, reflecting the nonrandom nature of GI insertion in P. aeruginosa (Wiehlmann et al. 2007). Significant sequence composition bias in seven of the nine regions was computationally identified (Table 2), while GC content deviating from the average can be observed for these regions in the figure.
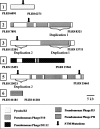
Figure 2.
Phage clusters identified in LESB58 with significant similarities and positioning of STM mutants after in vivo screening.
Figure 3.
Genomic islands identified in LESB58 with significant similarities and positioning of STM mutants after in vivo screening.
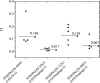
Figure 4.
In vivo competitive index (CI) of the P. aeruginosa STM PALES_45041 (within LESGI-5), PALES_25621 (within LES prophage 5), PALES_13261 (within LES prophage 3), and PALES_08021 (within LES prophage 2) grown for 7 d in the rat lung in competition with the wild-type LESB58 strain. Each circle represents the CI for a single animal in each group. A CI of less than 1 indicates an attenuation of virulence. The geometric mean of the CIs for all rats is shown as a solid line and statistically significant _P_-value is indicated with an asterisk (*P < 0.001 using the Mann-Whitney sum test).
Similar articles
- Differential infection properties of three inducible prophages from an epidemic strain of Pseudomonas aeruginosa.
James CE, Fothergill JL, Kade H, Hall AJ, Cottell J, Brockhurst MA, Winstanley C. James CE, et al. BMC Microbiol. 2012 Sep 21;12:216. doi: 10.1186/1471-2180-12-216. BMC Microbiol. 2012. PMID: 22998633 Free PMC article. - Genes Required for Free Phage Production are Essential for Pseudomonas aeruginosa Chronic Lung Infections.
Lemieux AA, Jeukens J, Kukavica-Ibrulj I, Fothergill JL, Boyle B, Laroche J, Tucker NP, Winstanley C, Levesque RC. Lemieux AA, et al. J Infect Dis. 2016 Feb 1;213(3):395-402. doi: 10.1093/infdis/jiv415. Epub 2015 Aug 12. J Infect Dis. 2016. PMID: 26268854 - Transmission, adaptation and geographical spread of the Pseudomonas aeruginosa Liverpool epidemic strain.
Moore MP, Lamont IL, Williams D, Paterson S, Kukavica-Ibrulj I, Tucker NP, Kenna DTD, Turton JF, Jeukens J, Freschi L, Wee BA, Loman NJ, Holden S, Manzoor S, Hawkey P, Southern KW, Walshaw MJ, Levesque RC, Fothergill JL, Winstanley C. Moore MP, et al. Microb Genom. 2021 Mar;7(3):mgen000511. doi: 10.1099/mgen.0.000511. Epub 2021 Mar 15. Microb Genom. 2021. PMID: 33720817 Free PMC article. - Phage Morons Play an Important Role in Pseudomonas aeruginosa Phenotypes.
Tsao YF, Taylor VL, Kala S, Bondy-Denomy J, Khan AN, Bona D, Cattoir V, Lory S, Davidson AR, Maxwell KL. Tsao YF, et al. J Bacteriol. 2018 Oct 23;200(22):e00189-18. doi: 10.1128/JB.00189-18. Print 2018 Nov 15. J Bacteriol. 2018. PMID: 30150232 Free PMC article. - Use of suppression subtractive hybridization to examine the accessory genome of the Liverpool cystic fibrosis epidemic strain of Pseudomonas aeruginosa.
Smart CHM, Walshaw MJ, Hart CA, Winstanley C. Smart CHM, et al. J Med Microbiol. 2006 Jun;55(Pt 6):677-688. doi: 10.1099/jmm.0.46461-0. J Med Microbiol. 2006. PMID: 16687584
Cited by
- Within-Host Evolution of the Dutch High-Prevalent Pseudomonas aeruginosa Clone ST406 during Chronic Colonization of a Patient with Cystic Fibrosis.
van Mansfeld R, de Been M, Paganelli F, Yang L, Bonten M, Willems R. van Mansfeld R, et al. PLoS One. 2016 Jun 23;11(6):e0158106. doi: 10.1371/journal.pone.0158106. eCollection 2016. PLoS One. 2016. PMID: 27337151 Free PMC article. - Identification of Novel Genomic Islands in Liverpool Epidemic Strain of Pseudomonas aeruginosa Using Segmentation and Clustering.
Jani M, Mathee K, Azad RK. Jani M, et al. Front Microbiol. 2016 Aug 3;7:1210. doi: 10.3389/fmicb.2016.01210. eCollection 2016. Front Microbiol. 2016. PMID: 27536294 Free PMC article. - What It Takes to Be a Pseudomonas aeruginosa? The Core Genome of the Opportunistic Pathogen Updated.
Valot B, Guyeux C, Rolland JY, Mazouzi K, Bertrand X, Hocquet D. Valot B, et al. PLoS One. 2015 May 11;10(5):e0126468. doi: 10.1371/journal.pone.0126468. eCollection 2015. PLoS One. 2015. PMID: 25961859 Free PMC article. - Pulmonary infection by Yersinia pestis rapidly establishes a permissive environment for microbial proliferation.
Price PA, Jin J, Goldman WE. Price PA, et al. Proc Natl Acad Sci U S A. 2012 Feb 21;109(8):3083-8. doi: 10.1073/pnas.1112729109. Epub 2012 Feb 1. Proc Natl Acad Sci U S A. 2012. PMID: 22308352 Free PMC article. - Filamentous prophage Pf4 promotes genetic exchange in Pseudomonas aeruginosa.
Pei TT, Luo H, Wang Y, Li H, Wang XY, Zhang YQ, An Y, Wu LL, Ma J, Liang X, Yan A, Yang L, Chen C, Dong T. Pei TT, et al. ISME J. 2024 Jan 8;18(1):wrad025. doi: 10.1093/ismejo/wrad025. ISME J. 2024. PMID: 38365255 Free PMC article.
References
- Akhverdian V.Z., Khrenova E.A., Bogush V.G., Gerasimova T.V., Kirsanov N.B. [Wide distribution of transposable phages in natural Pseudomonas aeruginosa populations] Genetika. 1984;20:1612–1619. - PubMed
- Bender C., Rangaswamy V., Loper J. Polyketide production by plant-associated Pseudomonads. Annu. Rev. Phytopathol. 1999;37:175–196. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases