Reference alignment of SNP microarray signals for copy number analysis of tumors - PubMed (original) (raw)
Reference alignment of SNP microarray signals for copy number analysis of tumors
Stan Pounds et al. Bioinformatics. 2009.
Abstract
A new procedure to align single nucleotide polymorphism (SNP) microarray signals for copy number analysis is proposed. For each individual array, this reference alignment procedure (RAP) uses a set of selected markers as internal references to direct the signal alignment. RAP aligns the signals so that each array has a similar signal distribution among its reference markers. An accompanying reference selection algorithm (RSA) uses genotype calls and initial signal intensities to choose two-copy markers as the internal references for each array. After RSA and RAP are applied, each array has a similar distribution of signals of two-copy markers so that across-array signal comparisons are biologically meaningful. An upper bound for a statistical metric of signal misalignment is derived and provides a theoretical basis to choose RSA-RAP over other alignment procedures for copy number analysis of cancers. In our study of acute lymphoblastic leukemia, RSA-RAP gives copy number analysis results that show substantially better concordance with cytogenetics than do two other alignment procedures.
Availability: Documented R code is freely available from www.stjuderesearch.org/depts/biostats/refnorm.
Figures
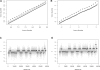
Fig. 1.
Alignment and segmentation results for a hyperdiploid tumor. (A) The quantiles of QA signals for markers on the amplified chromosomes (black dashed line) and chromosomes with no cytogenetic abnormality (solid black line) of a hyperdiploid tumor's array against the corresponding quantiles of QA signals for all markers of a female control tissue array. A diagonal gray line along _y_=x is included for reference. (C) The results of segmenting the standardized differences (
Supplementary Materials
, Section A) computed from QA signals. The _x_-axis represents markers (ordered by chromosome and position), and the _y_-axis represents the standardized differences. Each gray point represents the standardized difference for one marker, and the thick horizontal lines represent the median of the standardized differences in the determined segments. (B, D) panels show analogous results for the same tumor using RSA-RAP signals. RSA-RAP signals and QA signals are on different scales (A, B) because RSA-RAP maps signals to the targeted normal (0,1) distribution and QA maps to the empirically defined distribution of Equation (1).
Fig. 2.
ROC-type curves. (A–D) the concordance of each alignment procedure's results with cytogenetics across all tumor arrays as a function of the threshold γ among markers on amplified chromosomes, deleted chromosomes, chromosomes with no cytogenetically detected abnormality and all markers on chromosomes in one of these three categories. The solid black, dashed black, solid gray and dashed gray lines give the results for RSA-RAP, cyto-RAP, QA and ISA, respectively.
Similar articles
- SNiPer: improved SNP genotype calling for Affymetrix 10K GeneChip microarray data.
Huentelman MJ, Craig DW, Shieh AD, Corneveaux JJ, Hu-Lince D, Pearson JV, Stephan DA. Huentelman MJ, et al. BMC Genomics. 2005 Oct 31;6:149. doi: 10.1186/1471-2164-6-149. BMC Genomics. 2005. PMID: 16262895 Free PMC article. - A multi-array multi-SNP genotyping algorithm for Affymetrix SNP microarrays.
Xiao Y, Segal MR, Yang YH, Yeh RF. Xiao Y, et al. Bioinformatics. 2007 Jun 15;23(12):1459-67. doi: 10.1093/bioinformatics/btm131. Epub 2007 Apr 25. Bioinformatics. 2007. PMID: 17459966 - Implementation of array based whole-genome high-resolution technologies confirms the absence of secondary copy-number alterations in MLL-AF4-positive infant ALL patients.
Bardini M, Galbiati M, Lettieri A, Bungaro S, Gorletta TA, Biondi A, Cazzaniga G. Bardini M, et al. Leukemia. 2011 Jan;25(1):175-8. doi: 10.1038/leu.2010.232. Epub 2010 Oct 14. Leukemia. 2011. PMID: 20944671 No abstract available. - CGHcall: calling aberrations for array CGH tumor profiles.
van de Wiel MA, Kim KI, Vosse SJ, van Wieringen WN, Wilting SM, Ylstra B. van de Wiel MA, et al. Bioinformatics. 2007 Apr 1;23(7):892-4. doi: 10.1093/bioinformatics/btm030. Epub 2007 Jan 31. Bioinformatics. 2007. PMID: 17267432 - SNP array analysis in constitutional and cancer genome diagnostics--copy number variants, genotyping and quality control.
de Leeuw N, Hehir-Kwa JY, Simons A, Geurts van Kessel A, Smeets DF, Faas BH, Pfundt R. de Leeuw N, et al. Cytogenet Genome Res. 2011;135(3-4):212-21. doi: 10.1159/000331273. Epub 2011 Sep 16. Cytogenet Genome Res. 2011. PMID: 21934286 Review.
Cited by
- Analysis of the coding genome of diffuse large B-cell lymphoma.
Pasqualucci L, Trifonov V, Fabbri G, Ma J, Rossi D, Chiarenza A, Wells VA, Grunn A, Messina M, Elliot O, Chan J, Bhagat G, Chadburn A, Gaidano G, Mullighan CG, Rabadan R, Dalla-Favera R. Pasqualucci L, et al. Nat Genet. 2011 Jul 31;43(9):830-7. doi: 10.1038/ng.892. Nat Genet. 2011. PMID: 21804550 Free PMC article. - Significance of TP53 Mutation in Wilms Tumors with Diffuse Anaplasia: A Report from the Children's Oncology Group.
Ooms AH, Gadd S, Gerhard DS, Smith MA, Guidry Auvil JM, Meerzaman D, Chen QR, Hsu CH, Yan C, Nguyen C, Hu Y, Ma Y, Zong Z, Mungall AJ, Moore RA, Marra MA, Huff V, Dome JS, Chi YY, Tian J, Geller JI, Mullighan CG, Ma J, Wheeler DA, Hampton OA, Walz AL, van den Heuvel-Eibrink MM, de Krijger RR, Ross N, Gastier-Foster JM, Perlman EJ. Ooms AH, et al. Clin Cancer Res. 2016 Nov 15;22(22):5582-5591. doi: 10.1158/1078-0432.CCR-16-0985. Epub 2016 Oct 4. Clin Cancer Res. 2016. PMID: 27702824 Free PMC article. - The genetics of nodal marginal zone lymphoma.
Spina V, Khiabanian H, Messina M, Monti S, Cascione L, Bruscaggin A, Spaccarotella E, Holmes AB, Arcaini L, Lucioni M, Tabbò F, Zairis S, Diop F, Cerri M, Chiaretti S, Marasca R, Ponzoni M, Deaglio S, Ramponi A, Tiacci E, Pasqualucci L, Paulli M, Falini B, Inghirami G, Bertoni F, Foà R, Rabadan R, Gaidano G, Rossi D. Spina V, et al. Blood. 2016 Sep 8;128(10):1362-73. doi: 10.1182/blood-2016-02-696757. Epub 2016 Jun 22. Blood. 2016. PMID: 27335277 Free PMC article. - Mutations in CBL occur frequently in juvenile myelomonocytic leukemia.
Loh ML, Sakai DS, Flotho C, Kang M, Fliegauf M, Archambeault S, Mullighan CG, Chen L, Bergstraesser E, Bueso-Ramos CE, Emanuel PD, Hasle H, Issa JP, van den Heuvel-Eibrink MM, Locatelli F, Stary J, Trebo M, Wlodarski M, Zecca M, Shannon KM, Niemeyer CM. Loh ML, et al. Blood. 2009 Aug 27;114(9):1859-63. doi: 10.1182/blood-2009-01-198416. Epub 2009 Jul 1. Blood. 2009. PMID: 19571318 Free PMC article. - A procedure to statistically evaluate agreement of differential expression for cross-species genomics.
Pounds S, Gao CL, Johnson RA, Wright KD, Poppleton H, Finkelstein D, Leary SE, Gilbertson RJ. Pounds S, et al. Bioinformatics. 2011 Aug 1;27(15):2098-103. doi: 10.1093/bioinformatics/btr362. Epub 2011 Jun 22. Bioinformatics. 2011. PMID: 21697127 Free PMC article.
References
- Bolstad BM, et al. A comparison of normalization methods for high density oligonucleotide array based on variance and bias. Bioinformatics. 2003;19:185–193. - PubMed
- Mullighan CG, et al. Genes regulating B cell development are mutated in acute lymphoid leukaemia. Nature. 2007;446:758–764. - PubMed
- Pounds S, Cheng C. Statistical development and evaluation of gene expression data filters. J. Comput. Biol. 2005;12:482–495. - PubMed
- Raimondi SC, et al. Cytogenetics as a diagnostic aid for childhood hematologic disorders: conventional cytogenetic techniques, fluorescence in situ hybridization, and comparative genomic hybridization. In: Hanausek M, Walaszek Z, editors. Tumor Marker Protocols. Methods Molecular Medicine. Totowa, NJ: Humana Press; 1998. pp. 209–227. 1998.
Publication types
MeSH terms
Substances
Grants and funding
- R01 CA129541/CA/NCI NIH HHS/United States
- U01 GM061393/GM/NIGMS NIH HHS/United States
- U01GM61393/GM/NIGMS NIH HHS/United States
- R01 CA132946/CA/NCI NIH HHS/United States
- U01GM61374/GM/NIGMS NIH HHS/United States
- CA21765/CA/NCI NIH HHS/United States
- R01CA132946-01/CA/NCI NIH HHS/United States
- R01CA115422-01A1/CA/NCI NIH HHS/United States
- R01 CA115422/CA/NCI NIH HHS/United States
- R01CA129541-01/CA/NCI NIH HHS/United States
- P30 CA021765/CA/NCI NIH HHS/United States
- U01 GM061374/GM/NIGMS NIH HHS/United States