Characterizing heterogeneous cellular responses to perturbations - PubMed (original) (raw)
Characterizing heterogeneous cellular responses to perturbations
Michael D Slack et al. Proc Natl Acad Sci U S A. 2008.
Abstract
Cellular populations have been widely observed to respond heterogeneously to perturbation. However, interpreting the observed heterogeneity is an extremely challenging problem because of the complexity of possible cellular phenotypes, the large dimension of potential perturbations, and the lack of methods for separating meaningful biological information from noise. Here, we develop an image-based approach to characterize cellular phenotypes based on patterns of signaling marker colocalization. Heterogeneous cellular populations are characterized as mixtures of phenotypically distinct subpopulations, and responses to perturbations are summarized succinctly as probabilistic redistributions of these mixtures. We apply our method to characterize the heterogeneous responses of cancer cells to a panel of drugs. We find that cells treated with drugs of (dis-)similar mechanism exhibit (dis-)similar patterns of heterogeneity. Despite the observed phenotypic diversity of cells observed within our data, low-complexity models of heterogeneity were sufficient to distinguish most classes of drug mechanism. Our approach offers a computational framework for assessing the complexity of cellular heterogeneity, investigating the degree to which perturbations induce redistributions of a limited, but nontrivial, repertoire of underlying states and revealing functional significance contained within distinct patterns of heterogeneous responses.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Fig. 1.
Heterogeneous cellular populations can be characterized as mixtures of subpopulations with distinct phenotypes. For convenience of visualization, results are shown for k = 4 subpopulations. (A and B) Illustration of approach. (A) HeLa cells are treated with drugs, labeled with fluorescence markers, and imaged. Individual cellular regions are identified. (B) A high-dimensional vector of phenotypic features is extracted from each cell. Cells are assigned probabilities of belonging to subpopulations that have been identified by using a GMM. Scatter plot: cells are visualized as points via feature representation and PCA reduction to two dimensions; colored cell outlines colors in A and points in B correspond to most probable subpopulation assignment; ellipses represent unit standard deviation from the mean for each subpopulation. Shown are 3 representative cells taken from various locations in each ellipse. (C) Shown are subpopulation models for different marker sets. Models are built from “reference” sets of cells sampled from multiple conditions. Scatter plots are as in B. Stacked bar graphs represent (prior) probabilities of cell membership in each subpopulation. Sample cells from within 1 standard deviation of the mean phenotype are shown at right for each subpopulation (image colors correspond to markers in order; e.g., blue/green/red corresponds to DNA/SC35/anillin). (Scale bar: 20 μm.)
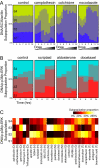
Fig. 2.
Subpopulation profiles allow quantitative comparison of heterogeneous cellular responses. (A) Subpopulation profiles reveal dose-dependent heterogeneous responses to drugs [DNA/SC35/anillin marker set, 20-h drug exposure (25)]. Thirteen concentrations are shown, increasing from left to right, at 3-fold titration. (B) Subpopulation profiles reveal time-dependent heterogeneous responses to drugs. Subpopulation profiles for selected drugs are combined in time series (DNA/_p_-p38/_p_-ERK marker set; concentrations are given in
Table S1
). Profiles in A and B are based on reference models computed with k = 4 subpopulations. Colors and subpopulations ids S1-S4 on left-most images correspond to Fig. 1_C_. (C) Drugs of similar mechanism often induce similar subpopulation profiles. Subpopulation profiles for 25 drugs at the 24-h time point and DNA/_p_-p38/_p_-ERK marker set are displayed in a heat map and grouped by category (D, DNA replication; H, histone deacetylase; M, microtubule; G, glucocorticoid receptor; T, topoisomerase). Drugs with <200 cells per well are marked with asterisks (mitomycin C and trifluoperazine).
Fig. 3.
A small number of phenotypic subpopulations can distinguish drug effects by mechanism. (A) As the number of subpopulations in the reference model increases (k = 2, 4, 10, and 100), MDS plots (40) reveal relative similarities among drugs (computed by using KL divergence) within 3 mechanism categories (H, blue diamonds; M, green triangles; G, orange stars). Axes are labeled in arbitrary units. (B) Drug classification performance is computed for each value of k (and drug category; see also
Table S5
). Performance is measured by −log10 of the P value for a rank-sum statistic [Mann–Whitney U test (39)] obtained by comparing distributions of intra- and intercategory subpopulation profile similarity scores. Profile similarity is computed for 12- and 24-h time points of low drug concentration and combined by rank addition. Dotted lines indicate significance threshold corresponding to P = 0.05. Black arrowheads (Left) (k = 2, 4, 10, 100) correspond to MDS plots in A. Black arrowheads (Right) (k = 7) correspond to MDS plot at bottom right and uses reference model shown in Fig. S1_D_.
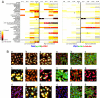
Fig. 4.
Exceptional phenotypes are detected by evaluating the fit of each drug to a reference model built by using only the low concentration of 1 drug in each functional category (control, aldosterone, aphidicolin, apicidin, colchicine, and doxorubicin) at all time points. (A) Reference models for DNA/_p_-p38/_p_-ERK and DNA/actin/α-tubulin marker sets are both computed with k = 4 subpopulations. Heat map reflects the ratio of model fit (log likelihood) of drug treatment data to training data. Four time points (3, 6, 12, and 24 h) are displayed for each of 2 marker sets, at both low and high concentrations of each drug. Gray boxes indicate poor fits due to low cell counts (<700 cells per well). (B) Shown are sample images of drug treated cells that fit the reference model well (a–c), as well as the 6 lowest scoring drugs (–6) as indicated in A.
Similar articles
- Patterns of basal signaling heterogeneity can distinguish cellular populations with different drug sensitivities.
Singh DK, Ku CJ, Wichaidit C, Steininger RJ 3rd, Wu LF, Altschuler SJ. Singh DK, et al. Mol Syst Biol. 2010 May 11;6:369. doi: 10.1038/msb.2010.22. Mol Syst Biol. 2010. PMID: 20461076 Free PMC article. - In vitro investigations of new therapeutic agents on bladder tumor cell lines.
Kugler A, Haschemi R, Zöller G, Gross AJ, Kallerhoff M, Ringert RH. Kugler A, et al. Urol Res. 1997;25(4):247-50. doi: 10.1007/BF00942093. Urol Res. 1997. PMID: 9286032 - Intratumor heterogeneity alters most effective drugs in designed combinations.
Zhao B, Hemann MT, Lauffenburger DA. Zhao B, et al. Proc Natl Acad Sci U S A. 2014 Jul 22;111(29):10773-8. doi: 10.1073/pnas.1323934111. Epub 2014 Jul 7. Proc Natl Acad Sci U S A. 2014. PMID: 25002493 Free PMC article. - [Anticancer drugs and pharmacologic actions].
Ogawa M. Ogawa M. Nihon Rinsho. 1997 May;55(5):1017-23. Nihon Rinsho. 1997. PMID: 9155146 Review. Japanese. - Tumour heterogeneity and metastasis at single-cell resolution.
Lawson DA, Kessenbrock K, Davis RT, Pervolarakis N, Werb Z. Lawson DA, et al. Nat Cell Biol. 2018 Dec;20(12):1349-1360. doi: 10.1038/s41556-018-0236-7. Epub 2018 Nov 26. Nat Cell Biol. 2018. PMID: 30482943 Free PMC article. Review.
Cited by
- Sphingolipid-Based Synergistic Interactions to Enhance Chemosensitivity in Lung Cancer Cells.
Mesén-Porras S, Rojas-Céspedes A, Molina-Mora JA, Vega-Baudrit J, Siles F, Quiros S, Mora-Rodríguez R. Mesén-Porras S, et al. Cells. 2023 Nov 8;12(22):2588. doi: 10.3390/cells12222588. Cells. 2023. PMID: 37998323 Free PMC article. - Patterns of basal signaling heterogeneity can distinguish cellular populations with different drug sensitivities.
Singh DK, Ku CJ, Wichaidit C, Steininger RJ 3rd, Wu LF, Altschuler SJ. Singh DK, et al. Mol Syst Biol. 2010 May 11;6:369. doi: 10.1038/msb.2010.22. Mol Syst Biol. 2010. PMID: 20461076 Free PMC article. - Stochastic sensitivity analysis and kernel inference via distributional data.
Li B, You L. Li B, et al. Biophys J. 2014 Sep 2;107(5):1247-1255. doi: 10.1016/j.bpj.2014.07.025. Biophys J. 2014. PMID: 25185560 Free PMC article. - In Vivo Autofluorescence Imaging of Tumor Heterogeneity in Response to Treatment.
Shah AT, Diggins KE, Walsh AJ, Irish JM, Skala MC. Shah AT, et al. Neoplasia. 2015 Dec;17(12):862-870. doi: 10.1016/j.neo.2015.11.006. Neoplasia. 2015. PMID: 26696368 Free PMC article. - Human nuclear hormone receptor activity contributes to malaria parasite liver stage development.
Mittal N, Davis C, McLean P, Calla J, Godinez-Macias KP, Gardner A, Healey D, Orjuela-Sanchez P, Ottilie S, Chong Y, Gibson C, Winzeler EA. Mittal N, et al. Cell Chem Biol. 2023 May 18;30(5):486-498.e7. doi: 10.1016/j.chembiol.2023.04.011. Epub 2023 May 11. Cell Chem Biol. 2023. PMID: 37172592 Free PMC article.
References
- Bahar R, et al. Increased cell-to-cell variation in gene expression in ageing mouse heart. Nature. 2006;441:1011–1014. - PubMed
- Bar-Even A, et al. Noise in protein expression scales with natural protein abundance. Nat Genet. 2006;38:636–643. - PubMed
- Colman-Lerner A, et al. Regulated cell-to-cell variation in a cell-fate decision system. Nature. 2005;437:699–706. - PubMed
- Ferrell JE, Jr, Machleder EM. The biochemical basis of an all-or-none cell fate switch in Xenopus oocytes. Science. 1998;280:895–898. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM081549/GM/NIGMS NIH HHS/United States
- K22 CA118717/CA/NCI NIH HHS/United States
- R01 GM071794/GM/NIGMS NIH HHS/United States
- P50 CA070907/CA/NCI NIH HHS/United States
- K22 CA118717-01/CA/NCI NIH HHS/United States
- P50 CA70907/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources